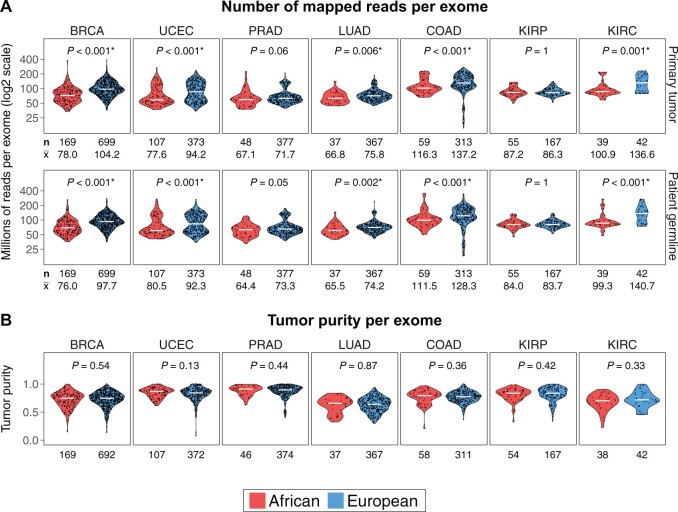
Figure 1.
Comparison of exome sequencing depths between ancestrally European and African patients in 7 The Cancer Genome Atlas cancer types that each profiled at least 50 self-reported Black patients. A) Number of mapped reads per exome for primary tumor (upper panel) and patient-matched germline (lower panel). Mann-Whitney U test 2-sided P values are shown for all comparisons. Sequencing depths of both tumor and germline exomes from ancestrally European patients were statistically significantly higher than those of ancestrally African patients in breast cancer (BRCA; breast invasive carcinoma), LUAD (lung adenocarcinoma), UCEC (uterine corpus endometrial carcinoma), KIRC (kidney renal clear cell carcinoma), and COAD (colon adenocarcinoma). The number of patients in either ancestral group (n) and the average sequencing depth per sample in millions of reads () are displayed below the box plots. B) There were no differences in tumor purity between ancestrally African and European tumors in all 7 cancer types. The numbers of patients with available tumor purity data in either ancestral group are displayed below the box plots. KIRP = kidney renal papillary cell carcinoma; PRAD = prostate adenocarcinoma.