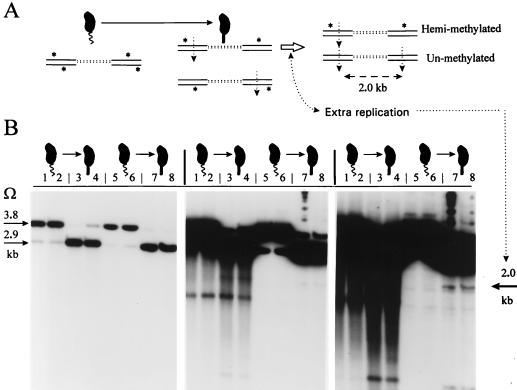
FIG. 7.
Sensitive detection of extra chromosome replication by assaying unmethylated DNA in stalked cells. (A) Rationale for assay. Swarmer cells differentiate into stalked cells, producing hemimethylated Tn5Ω-MP DNA, drawn as paired double lines where top or bottom asterisks denote GANTC methylation and vertical arrows denote susceptible ClaI or XhoI endonuclease sites (compare Fig. 2A). The open horizontal arrow indicates that one of the two replicating stalked-cell chromosomes has started a second round of replication, and in the absence of CcrM, this will produce sister hemimethylated and unmethylated DNA duplexes. The absence of methylation allows simultaneous left and right cleavage of the same Tn5Ω-MP, producing the 2.0-kb band (compare Fig. 2A). (B) Potentially unmethylated 2.0-kb chromosome DNA band, revealed during progressive 1- and 18-h and 4-day Southern blot exposures of the following experiment. DNA was prepared from the synchronous culture of Fig. 6 for T = 0 and T = 10 min (lanes 1 and 5, and 2 and 6) swarmer cells and from T = 50 and T = 63 min (lanes 3 and 7, and 4 and 8) stalked cells. Both standard phenol-chloroform extraction (lanes 1 to 4) and the agarose-embedding (lanes 5 to 8) methods for preparing DNA are compared. The Southern blotting protocol was performed as detailed in Materials and Methods and interpreted in Fig. 3A. DNA in each of lanes 1 to 8 was cut with ClaI and PstI, and the resulting blot was hybridized with >3 × 109 cpm of 32P-labeled Ω fragment/μg (Fig. 3A). Size standards, in the adjacent lanes (not shown), were the 1-kb ladder (Gibco BRL) and blotted chromosome DNA cut with HindIII, PstI, and XhoI. The arrows mark the 3.8-, 2.9-, and 2.0-kb bands corresponding to fully methylated, hemimethylated, and unmethylated DNAs (Fig. 3A).