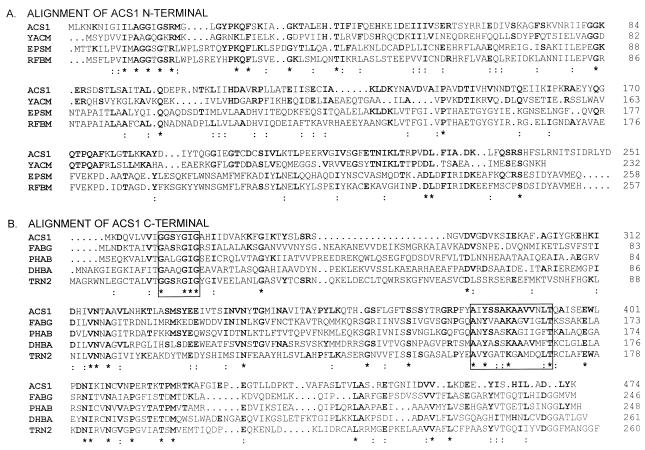
FIG. 3.
Alignment of the predicted amino acid sequence of Acs1 with homologous proteins detected by BLAST and FASTA searches. Multiple sequence alignments were performed with the CLUSTAL W program (45). Conserved residues are in boldface; ∗, identical residues; :, amino acids belonging to the same physicochemical group. (A) Alignment of Acs1 amino acids 1 to 251. YACM, B. subtilis hypothetical 25.8-kDa enzyme, unidentified protein family UPF0007 (33% identity) (35); EpsM, Acinetobacter calcoaceticus bifunctional phosphomannose-isomerase–GDP-mannose-pyrophosphorylase (unpublished; TrEMBL accession no. Q43941); RFBM, Salmonella typhimurium mannose 1-P-guanylyltransferase (25). (B) Alignment of Acs1 amino acids 252 to 474. FABG, B. subtilis 3-oxoacyl acylcarrier protein reductase (30); PHAB, acetoacetyl-CoA reductase from Acinetobacter sp. strain RA3849 (42); DHBA, B. subtilis 2,3-dihydro-2,3-dihydroxylbenzoate dehydrogenase (1); TRN2, Hyoscyamus niger tropinone reductase II (33). All homologous proteins are members of the SDR family. The NAD(P) binding site is indicated by a box at amino acids 261 to 267. The sequence motif typical of the SDR family (18) is contained in a box at amino acids 380 to 393.