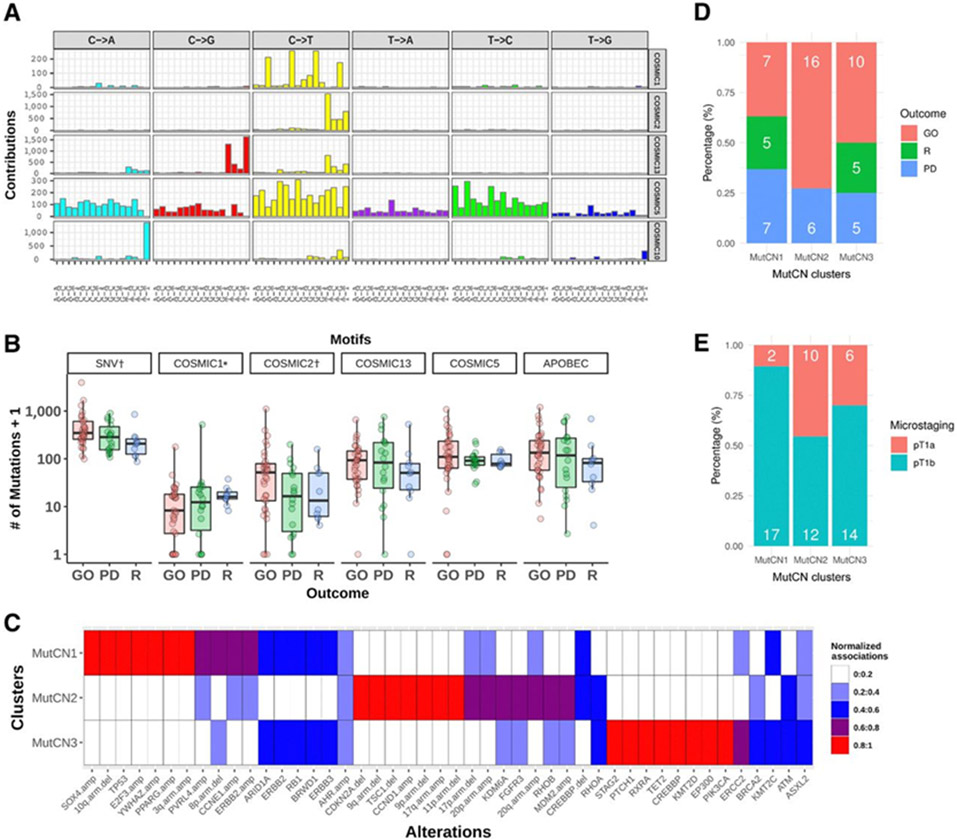
Figure 3.
A, Mutational signatures of 62 high-grade non–muscle-invasive urothelial tumors identified by a Bayesian nonnegative matrix factorization algorithm (top to bottom). Five distinct mutational signatures were identified that are most similar to COSMIC signatures 1, 2, 5, 10, and 13 (17). Two are due to APOBEC activity (COSMIC2, COSMIC13; APOBEC-A and APOBEC-B, respectively); one is attributable to POLE mutation (COSMIC10); one more uniform mutation signature that resembles COSMIC signature 5; and finally a signature characterized by C>T at CpG that corresponds to the aging signature (COSMIC1). However, two samples in which this signature was predominant (vh122 and vh73, Supplementary Fig. S4A) are MSI high samples, and hence in those two samples, the signature likely reflects the mutagenic effects of defective DNA mismatch repair. B, Association of mutational signatures and SNVs with disease outcome (excluding POLE sample vh102). *, COSMIC1 includes the two samples dominated by COSMIC6 signature. Significant associations (P < 0.05) are indicated by †. Y axis is exponential scale. C, Mutation and focal CN (MutCN) clusters. Normalized strength of association of 46 genetic alterations to the three MutCN clusters is shown. D, Frequency of disease outcome. E, Microstaging by MutCN clusters. Barplots depict the percentage of patients (number of patients in white).