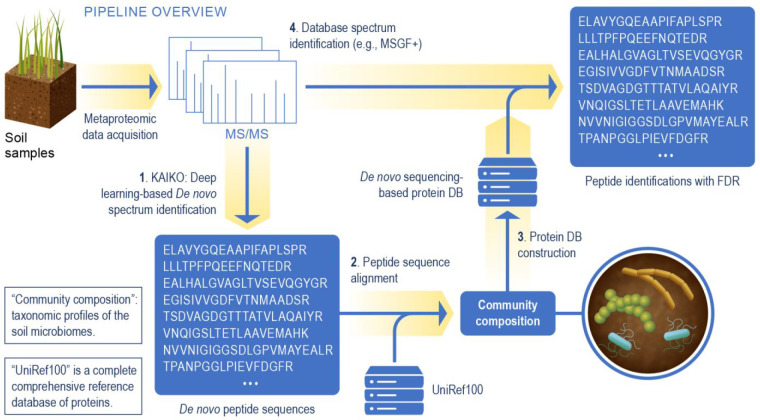
Figure 2.
Overview of the metaproteomics data analysis leveraging de novo spectrum identification based on the Kaiko model. Peptides are identified using Kaiko and used to infer community composition (steps 1–3). In step 4, the spectra are reanalyzed using a database search algorithm, e.g., MSGF+, and the protein sequence database created in step 3. This yields a final list of peptide identifications which can be used for functional analysis.