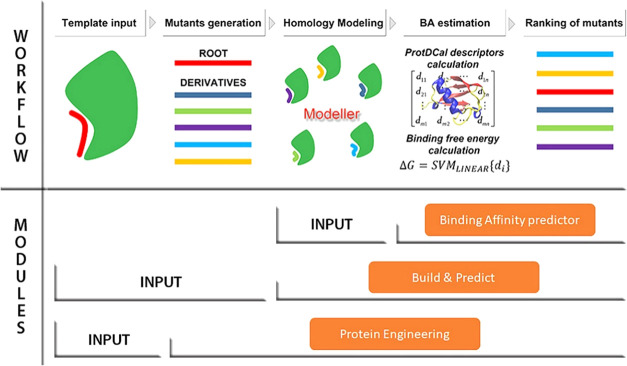
Figure 5.
Workflow of the three modules included in PPI-Affinity. For each module, the required input data and associated steps are indicated. The main difference among them lies in the input data: the Binding Affinity predictor module receives as input a set of protein–protein/peptide complexes (in PDB format); the Build & Predict module generates the complexes from a template file and a list of amino acid sequences (in FASTA format) provided by the user; and the Protein Engineering module receives as input only a template model, generates a list of derivatives for one of the protein/peptide contained in the PDB file, and calculates homology models for all created mutants. Regardless of the module, once the PDB files have been prepared, PPI-Affinity computes the structural descriptors with ProtDCal, estimates the BA values with the machine learning models, and returns the list of derivatives ranked by their BA values.