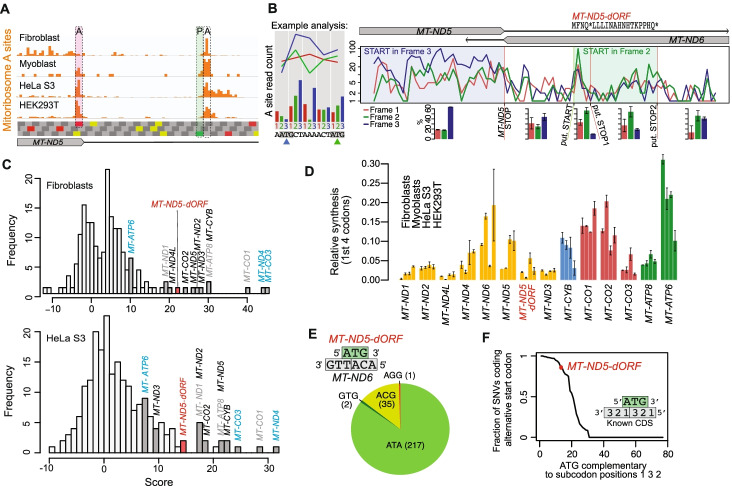
Fig. 2.
Evidence for the translation of a novel mitochondrial noncanonical open reading frame. A Translation initiation occurs on an internal AUG in the MT-ND5 3′ UTR across cell lines. A-site density on the MT-ND5 stop codon (shaded in red) and P-site density (shaded in green, A-site density one codon downstream) of initiating ribosomes on the internal AUG are highlighted. Codons in the three frames are shown below with stop (UAA, UAG), start (AUG), and mitochondrial alternative start (AUU, AUA) highlighted (red, green, yellow, respectively). B Stacked frame plot highlighting number of A-site transformed reads in each subcodon position on heavy strand. The example analysis (left panel) shows how raw A site read counts are transformed to line plots, and how the line plots highlight which frame is being read. The color of the arrowheads indicates which frame each ATG is in. The blue ATG starts an open reading frame. Mitoribosome profiling data are from two fibroblast replicates, summed. Amino acid sequence of putative MT-ND5-dORF peptide is shown with putative (put.) start and stop codons highlighted with green and red lines, respectively. Bar plots below show the average percentage of reads in each subcodon position for regions indicated, with error bars showing range between the replicates summed above. C Distribution of initiation scores after applying ad hoc scoring method to fibroblasts and HeLa S3 cells. Gene names in black: known genes with no leader; gray: known genes with 1 to 3 nt leader; cyan: known genes with long 5′ UTR. D Estimated relative synthesis of MT-ND5-dORF compared to other mtDNA-encoded genes. A-site transformed reads were summed across the first four codons with signal for each gene and normalized by the total number of such reads. For genes with 5′ UTRs (MT-ATP6, MT-ND4, MT-CO3, MT-ND5-dORF), this includes codons 2–5, where codon 5 is a stop codon for MT-ND5-dORF. For genes with a very short or no leader (all others), this includes codons 6–9. For MT-ATP6, the signal is confounded by reads from MT-ATP8, which overlaps the first 15 codons. Samples are plotted in the order listed and error bars show range across replicates. E Variants at MT-ND5-dORF ATG (in green), which is antisense to MT-ND6 (in grey) in the orientation indicated by the schematic. ATA: canonical alternative mitochondrial start codon. GTG: variant start codon shown to efficiently initiate translation. ACG: capable of initiation in reconstituted mammalian mitochondria translation (Lee et al, 2021). AGG: ambiguous (stop or frame-shift inducing). F Genome-wide analysis of SNVs at ATGs. Only equivalently oriented ATGs opposite to codons in known ORFs were included to equalize the constraints (see “Methods”). ATGs were required to have at least 2 SNVs to be included