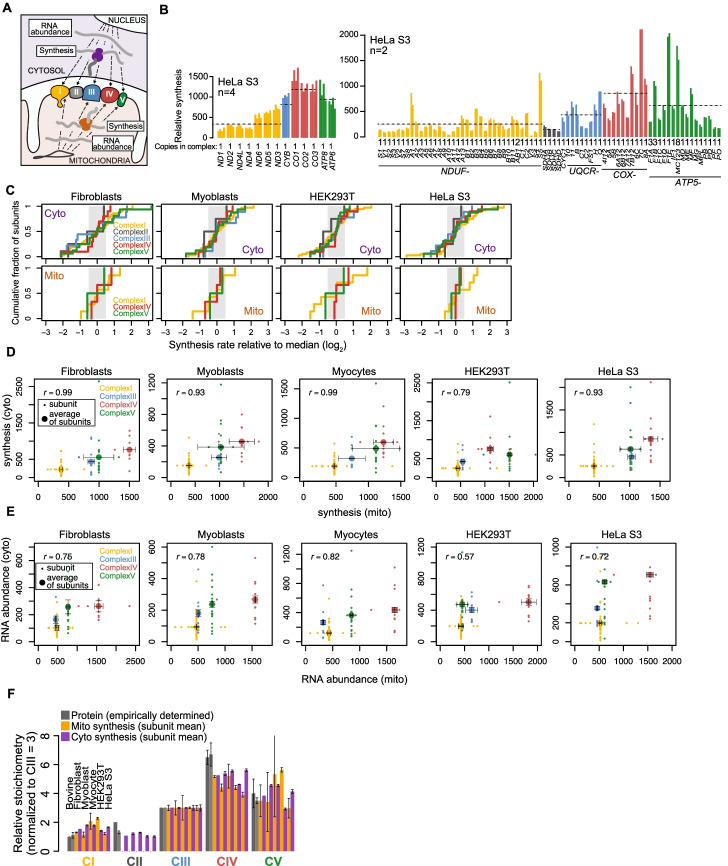
Fig. 3.
Balanced translatomes exist in five human cell lines. A Flow of genetic information to encode dual-origin OXPHOS complexes in human cells. B Relative synthesis values of mtDNA- (left graph) and nDNA-encoded (right graph) OXPHOS subunits. Cytoribosome profiling data are from Wu et al. [42]. Each replicate is shown as an individual bar, values given in Additional file 3: Table S2. Dotted lines show averages across all subunits and samples. Numbers below bars show stoichiometry of each subunit. Synthesis units are tpm/100 for mitochondrial synthesis and tpm for cytosolic synthesis (note that mitochondrial and cytosolic synthesis absolute values cannot be compared across compartments). C Cumulative distribution of relative synthesis rates for nDNA-encoded subunits (top, Additional file 3: Table S2 and “Methods” for details) and mtDNA-encoded subunits (bottom) of the dual-origin OXPHOS complexes. Log2-fold differences in synthesis levels compared to the median value for each complex are shown. Shaded region indicates 2-fold spread. D Synthesis of mtDNA-encoded subunits (tpm/100) compared to synthesis of nDNA-encoded core subunits (tpm) for each complex. Small dots show individual subunits, large dots show average synthesis of subunits within each corresponding complex, following the color code displayed. Fibroblast and HeLa S3 cytoribosome profiling data are from Tirosh et al. [43] and Wu et al. [42], respectively. Myocytes are post-differentiation-induction day 2 myoblasts. Individual subunit synthesis for each replicate is shown in Fig. 3B (HeLa S3), and Additional file 1: Figure S3A. Error bars show range in averages across replicates. E RNA abundance of mtDNA-encoded subunits (tpm/100) compared to RNA abundance of nDNA-encoded subunits (tpm) for each complex. Subunits included are identical to those in D. Small dots show individual subunit RNA abundance, large dots show average RNA abundance of subunits within each corresponding complex, following the color code displayed. Error bars show range in averages across replicates. All panels show Pearson correlation, r. F Stoichiometry of average synthesis values for each complex relative to Complex III, which is set to 3, following published convention. Dark gray and light gray bars show empirically determined complex stoichiometry as determined in [1, 2], respectively