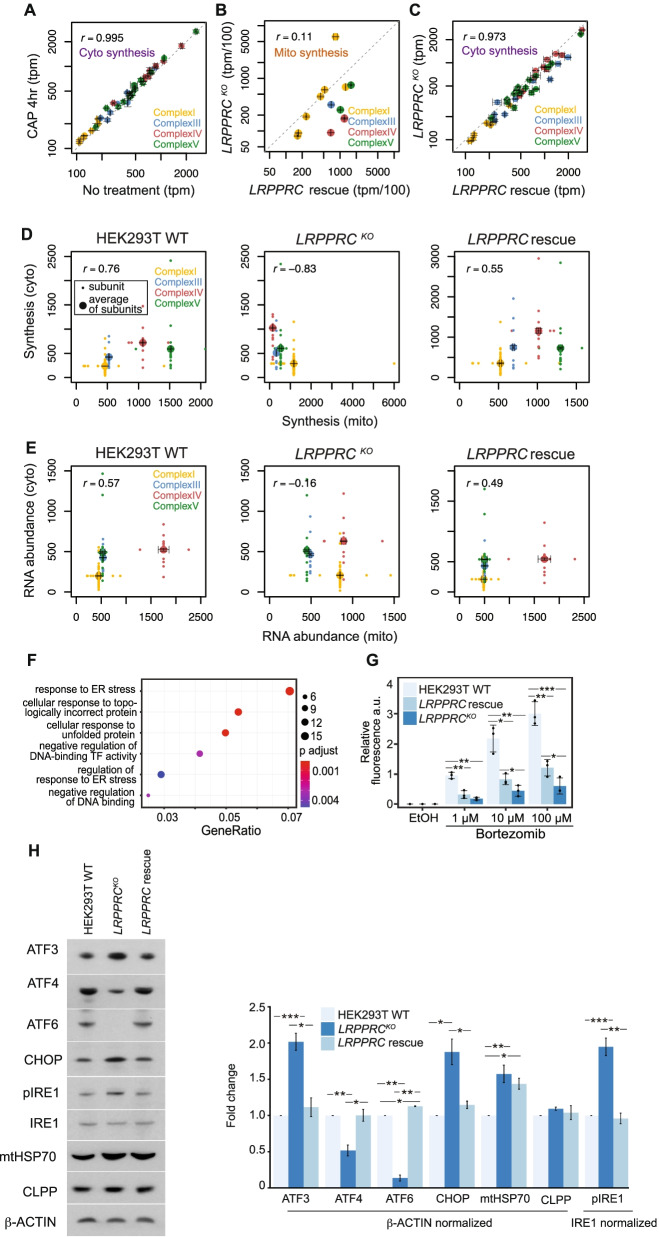
Fig. 4.
Lack of feedback between translatomes results in their imbalance and leads to the activation of proteostasis genes. A Comparison of relative synthesis levels for cytosolic-translated OXPHOS subunits with and without 4 h of chloramphenicol (CAP) treatment (100 μg/mL) to inhibit mitochondrial translation. B Relative synthesis for mitochondrial-encoded OXPHOS subunits in LRPPRCKO cells compared to a reconstituted cell line (LRPPRC rescue), expressing LRPPRC in the LRPPRCKO background. C Relative synthesis comparison as in (B) but for nuclear-encoded OXPHOS subunits. D Average synthesis of mtDNA-encoded subunits (tpm/100) compared to average synthesis of nDNA-encoded core subunits (tpm) for each complex in wild-type HEK293T cells, LRPPRCKO cells, and LRPPRC rescue cells. E Average RNA abundance of mtDNA-encoded subunits (tpm/100) compared to average RNA abundance of nDNA-encoded core subunits (tpm) for each complex in LRPPRC cell lines. Error bars throughout show range in averages across replicates, and all panels show Pearson correlation, r. F GO term enrichment analysis of significantly (adjusted p-value < 0.05) differentially expressed genes that show a minimum twofold increase in the LRPPRCKO cells compared to the LRPPRC rescue cell line. Shown are the GO terms after filtering for redundancy. The size of points represents the number of genes in each GO term. p adjust = adjusted p-value. TF = transcription factor. G Cell toxicity graph after 72-h bortezomib treatment. HEK293T WT, LRPPRCKO, or LRPPRC rescue cells were treated with increasing amounts of bortezomib or the vehicle ethanol (EtOH). Cell death was measured via a fluorescence dye and living cells by indirect measurement of ATP levels via luminescence. Shown are the mean relative fluorescence arbitrary units (a.u.) normalized to the respective luminescence of 3 technical replicates, with WT ethanol treated having 2 technical replicates. The range bars represent the standard deviation. The stars represent significant p-values, determined with a Welch two-sample one-sided (less) t test, * signifies p-value < 0.05, ** signifies p-value < 0.01 and *** signifies p-value < 0.001. Two other biological replicates are shown in Additional file 1: Figure S4L. H Left panel shows the immunoblot analysis using whole cell lysates from HEK293 WT, LRPPRCKO, or LRPPRC rescue cells probed for antibodies of markers of the ER and mitochondrial UPR pathways. Bar graphs show signals normalized by β-ACTIN values and compared to WT to determine fold changes. IRE1 levels are shown as ratios of its phosphorylated form (pIRE1) compared to total (IRE1). Error bars indicate standard deviation between two replicates. T test was used to test for significant changes. * signifies p-value < 0.05, ** signifies p-value < 0.01, and *** signifies p-value < 0.001