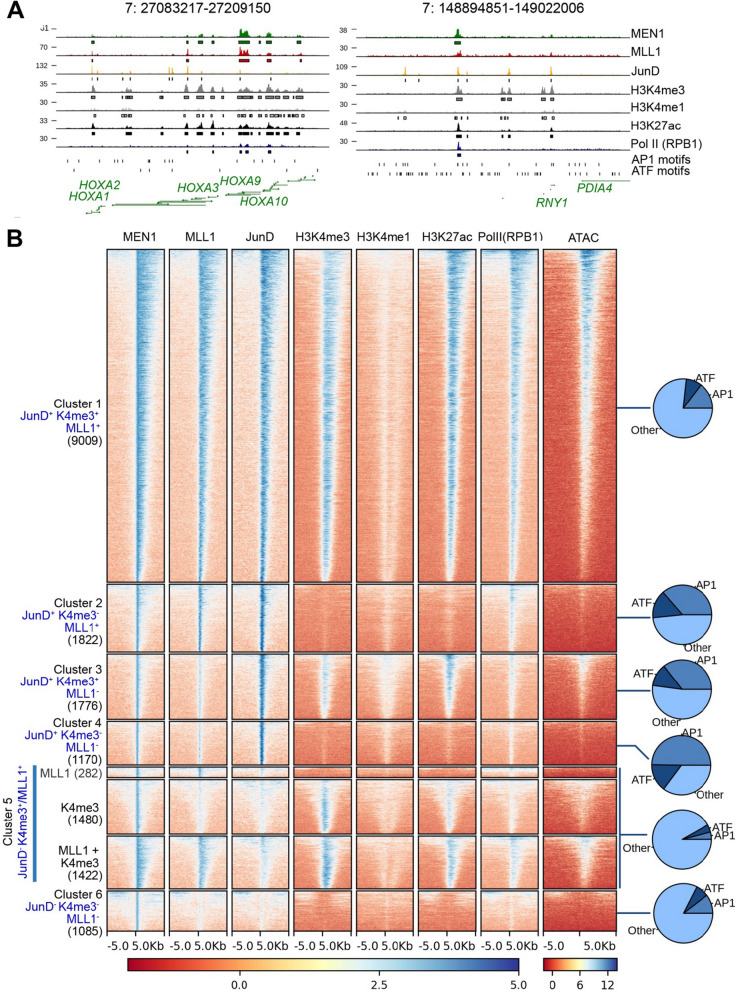
Fig. 5.
Genome-wide occupancy of WT menin, MLL1, JunD, RNA polymerase II, genome-wide presence of histone marks and DNA accessibility in HeLa cells. A Normalized coverage tracks of the menin occupied regions showing broad and narrow peaks. B Normalized heatmaps indicating menin, MLL1, JunD, H3K4me3, H3K4me1, H3K27ac and pol II peaks, as well as ATACseq results categorized according to menin, MLL1 and JunD binding. The + and – signs indicate overlap and non-overlap with the peaks of WT menin, respectively. The eight categories spanning six clusters are indicated on the left and the numbers between brackets indicate the total number of menin peaks in each cluster. Pie charts showing the proportion of AP-1 and ATF motifs at menin–JunD sites in the different clusters are shown on the right side of heatmaps. Other indicates the proportion of peaks without AP-1 and ATF motifs