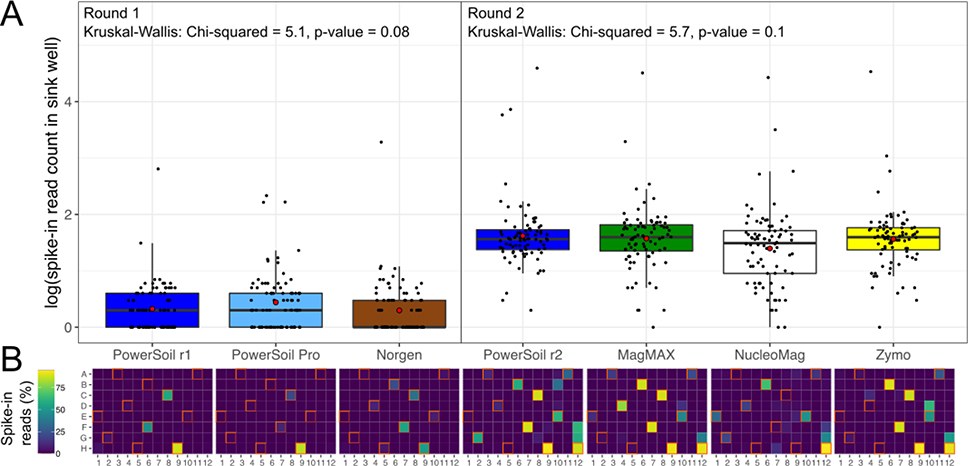
Figure 3.
Well-to-well contamination across candidate extraction kits as compared to our standardized protocol. Plasmids harboring synthetic 16S sequences were spiked into a single well per plate column (i.e., 1–11) of each high-biomass sample plate prior to extraction. (A) The number of reads matching synthetic 16S sequences was quantified for all wells that did not receive a spike-in (i.e., sink wells). Round 1 and Round 2 indicate different sequencing runs, and because sampling effort was not normalized here such to compare absolute read counts, comparisons should not be made across sequencing runs. For each sequencing run, results from a Kruskal-Wallis test are shown. (B) The percentage of spike-in reads among all reads per well shown as a heatmap. Wells into which plasmids were spiked (i.e., source wells) are outlined in orange.