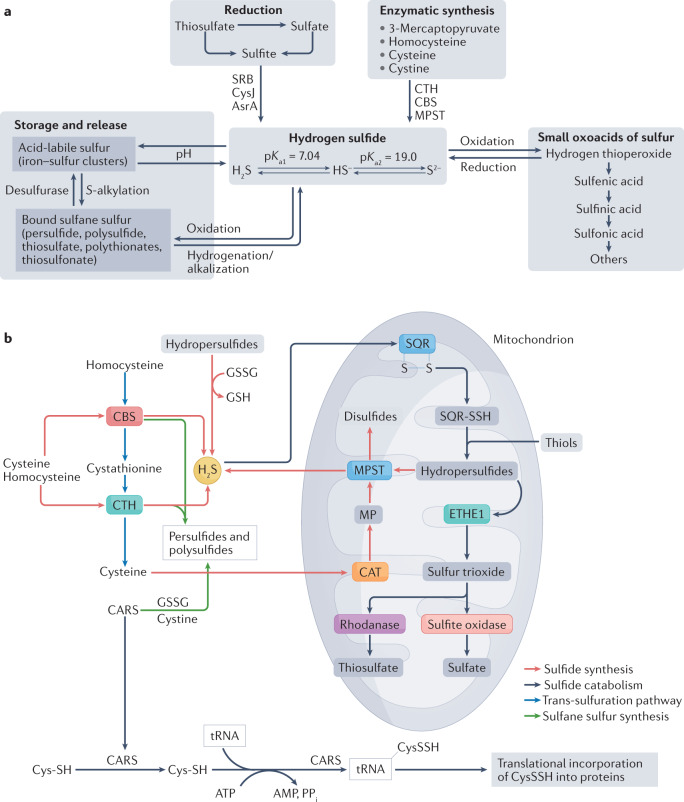
Fig. 1. Sulfide metabolite formation and fate.
a | Various chemical metabolite fate pathways for sulfide and its related species are shown. The basal level of production of hydrogen sulfide (H2S) is determined by the activity of three main enzymes: cystathionine γ-lyase (CTH), cystathionine β-synthase (CBS) and 3-mercaptopyruvate sulfurtransferase (MPST). In addition, bacterial enzymes (such as sulfate-reducing bacteria (SRB), sulfite reductase [NADPH] flavoprotein α-component (CysJ) and anaerobic sulfite reductase subunit A (AsrA)) can reduce terminal sulfide oxidation end products (such as thiosulfate, sulfate and sulfite) back to H2S. H2S can undergo a myriad of reactions leading to the formation of small oxoacids of sulfur, sulfane sulfur species and acid-labile sulfur species. b | Various enzymatic and non-enzymatic biochemical pathways are involved in sulfide metabolite formation. Sulfide catabolism through the mitochondrial H2S oxidation pathway leads to the metabolic end products of sulfate and thiosulfate. CARS, cysteinyl–tRNA synthetase (also known as cytoplasmic cysteine–tRNA ligase); CAT, cysteine aminotransferase; CysSH, cysteine; CysSSH, cysteine hydropersulfide; ETHE1, persulfide dioxygenase; GSH, glutathione; GSSG, glutathione disulfide; MP, mercaptopyruvate; PPi, inorganic pyrophosphate; SQR, sulfide–quinone oxidoreductase; SQR-SSH, sulfide–quinone oxidoreductase hydropersulfide.