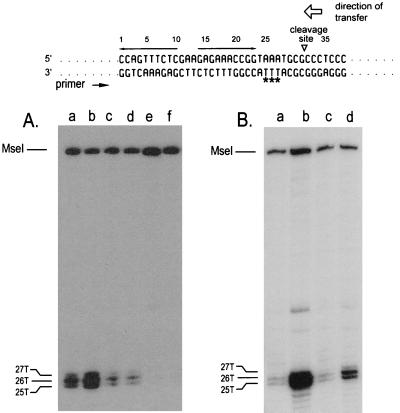
FIG. 4.
(A) Permanganate-sensitive bases on the unnicked oriT DNA strand for pUT1530 (lanes a and b), pUT1562 (lanes c and d), and pUT1532 (lanes e and f). The locations of the bases were determined from a sequencing ladder (not shown) generated with the same template and primer. Plasmid DNAs in cleared lysates were treated with permanganate prior to primer extension by PCR (31, 32). The cells also contained either pACYC184 (lanes a, c, and e) or pUT221 (lanes b, d, and f). (B) Permanganate sensitivity of the unnicked oriT DNA strand for pUT1530 isolated from cells also containing pBR322 (lane c) or pUT530 (lane d). Whole cells were exposed to permanganate prior to extraction of the DNA (31). For comparison, the permanganate sensitivity of pUT1530 DNA isolated from cells treated under identical conditions but containing pACYC184 (lane a) or pUT221 (lane b) is also shown. The base sequence of oriT is given at the top of the figure; the permanganate-sensitive bases are identified by asterisks. The inverted repeat (opposing arrows), the site cleaved by MobA in the top (transferred) strand, and the direction of transfer are also indicated.