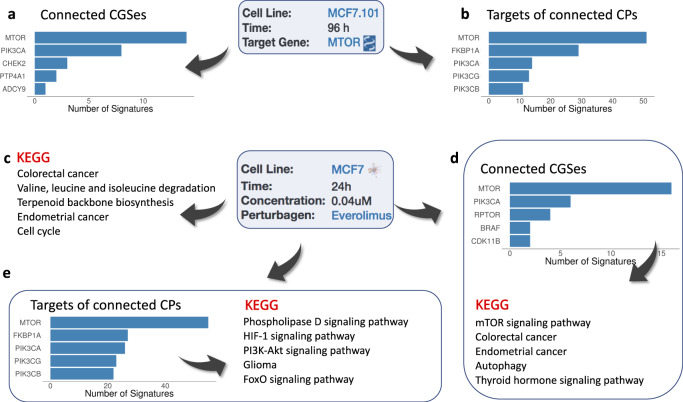
Fig. 2. Analysis of LINCS L1000 signatures of genetic and chemical perturbations.
a Most frequently perturbed genes among the Consensus Genes Signatures (CGS) connected to the mTOR knockdown CGS. b Most frequent inhibition targets of chemical perturbagens with signatures connected to the mTOR CGS signature. c Most enriched biological pathways for the everolimus signature. d Most frequently perturbed genes among CGSes connected with everolimus signature, and pathways most enriched by the perturbed genes. e Most frequent inhibition targets of chemical perturbagens with signatures connected to the everolimus signature and the pathways most enriched by the genes of the targeted proteins.