Abstract
Healthy pregnancy requires tolerance to fetal alloantigens as well as syngeneic embryonic and placental antigens. Given the importance of the autoimmune regulator (Aire) gene in self-tolerance, we investigated the role of Aire-expressing cells in maternal-fetal tolerance. We report that maternal ablation of Aire-expressing (Aire+) cells during early mouse pregnancy caused intrauterine growth restriction (IUGR) in both allogeneic and syngeneic pregnancies. This phenotype is immune-mediated, as IUGR was rescued in Rag1-deficient mice, and involved a memory response, demonstrated by recurrence of severe IUGR in second pregnancies. Single-cell RNA sequencing demonstrated that Aire+ cell depletion in pregnancy results in expansion of activated T cells, particularly T follicular helper cells. Surprisingly, selective ablation of either Aire-expressing medullary thymic epithelial cells (mTECs) or extrathymic Aire-expressing cells (eTACs) mapped the IUGR phenotype exclusively to eTACs. Thus, we report a previously undescribed mechanism for the maintenance of maternal-fetal immune homeostasis and demonstrate that eTACs protect the conceptus from immune-mediated IUGR.
One Sentence Summary:
Maternal extrathymic Aire-expressing cells support maternal-fetal tolerance and prevent intrauterine growth restriction in mice.
Introduction
Healthy pregnancy requires maternal tolerance to antigens expressed by the conceptus. In addition to paternal antigens which are allogeneic to the mother, there are also multiple placental and embryonic antigens encoded in the maternal genome, produced when the mother herself was a fetus. Such pregnancy-associated antigens (PAAs) are therefore technically “self” antigens for the mother, but in postnatal life, are only encountered in the context of pregnancy. Whether maternal self-tolerance to PAAs protects pregnancy is an important biological question. Early pregnancy loss and intrauterine growth restriction (IUGR) are significant health problems worldwide and while there is a clinical association between autoimmunity and these pregnancy complications (1, 2) the immune mechanisms of this association have not been described.
The Autoimmune Regulator gene (Aire) plays an essential role in preventing autoimmunity by facilitating expression of tissue-restricted self-antigens in medullary thymic epithelial cells (mTECs), leading to clonal deletion (3) or regulatory conversion (4) of self-reactive T cells. Extrathymic Aire-expressing cells (eTACs), myeloid antigen-presenting cells in the secondary lymphoid organs, may also play a role in the maintenance of immune tolerance and further prevent autoimmunity (5, 6). Notably, Aire-regulated tissue-restricted antigens include placental antigens in mouse and human mTECs (7) and thymic Aire-expression is sexually dimorphic in reproductive-aged mice and humans (8, 9). Roles for Aire in decidualization and embryo implantation have also been proposed (10, 11, 12). Although patients with AIRE mutations on appropriate hormone replacement therapy can achieve pregnancy, these are nonetheless complicated by fetoplacental insufficiency, IUGR, and placentas with diffuse lymphocytic infiltration of the decidual plate, suggestive of placental autoimmunity (13, 14). However, whether Aire-expressing cells are functionally involved in maternal-fetal tolerance has not been investigated.
To study the role of Aire+ cells in maternal-fetal tolerance, we used transgenic Aire diphtheria toxin receptor (Aire-DTR) mice, in which Aire-expressing cell populations can be deleted by injection of diphtheria toxin (DT) (15); this model allowed us to avoid the potential confounders of chronic endocrine organ autoimmunity seen in constitutive Aire-deficient mice that can affect hormonal cycling and oocyte quality before pregnancy (11, 16, 17). We thus depleted Aire+ cells selectively during early pregnancy, because maternal adaptive immune tolerance of the fetus has been shown to be essential in this period in both syngeneic and allogeneic settings (18, 19).
Results
Aire-expressing cell depletion in allogeneic and syngeneic pregnancies
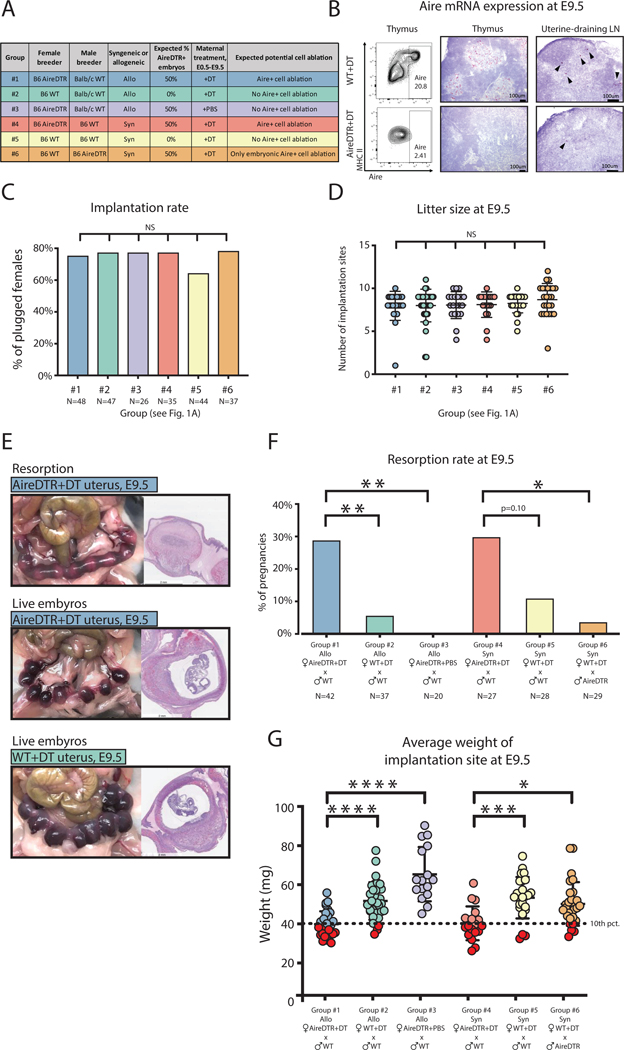
In order to ablate maternal Aire+ cells in allogeneic and syngeneic pregnancies, and control for off-target effects of DT and the AireDTR transgene, we used a combination of breeding approaches (Fig. 1A). We confirmed mTEC and eTAC depletion using flow cytometry and in situ hybridization (Fig. 1B). Implantation rates (Fig. 1C) and litter size did not differ significantly among experimental groups (Fig. 1D), indicating a post-implantation phenotype.
Figure 1. Maternal Aire+ cell ablation during pregnancy increases risk of resorption and intrauterine growth restriction.
(A) Experimental design: B6.AireDTR or B6.WT females were bred as indicated and treated with DT or PBS every other day from the day of plug (E0.5) until harvest (E9.5). (B) Flow cytometry of maternal thymi gated on CD11c- CD45- EpCAM+ mTECs (left), and in situ hybridization for Aire transcript (red) on maternal thymus (middle) and uterine-draining lymph nodes (right) of WT and AireDTR females treated with DT. (C) Implantation rate (percentage of plugged females with visible implantation sites at E9.5). N=number of plugged females. (D) Litter size at E9.5. (E) Gross pathology and H&E stain of pregnancy outcomes at E9.5: (top) AireDTR+DT uterus with resorbing embryos; (middle) AireDTR+DT uterus with live embryos; (bottom) WT+DT uterus with live embryos. (F) Resorption rate at E9.5. N=number of implanted pregnancies. (G) Average weight of implantation site (uterus weight / number of embryos). Dotted line = 10th percentile of implantation weight in the allogeneic WT+DT group. Red circles indicate concepti below the 10th percentile. ns = not statistically significant, p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 by Fisher’s exact test for (C) and (F); by Tukey’s multiple comparison test for (D) and (G).
Notably, we detected a significant increase in resorption (fetal wastage) rate after Aire+ cell depletion in allogeneic pregnancies compared to non-depleted allogeneic pregnancies (Fig. 1E, F). Further, we detected fetal growth restriction even in pregnancies without resorption: the average weight of implantation sites was significantly lower in the AireDTR+DT allogeneic pregnancies (group #1) than in the WT+DT allogeneic controls (group #2) (Fig. 1E, G). More than half of AireDTR+DT pregnancies had an average implantation site weight less than the 10th percentile of the WT+DT group, and met criteria for IUGR (20). This was not a transgene-intrinsic effect, as AireDTR+PBS allogeneic pregnancies (group #3) had pregnancy outcomes and implantation site weights comparable to those of WT+DT dams (Fig. 1F, G).
To determine whether these effects were due to the expression of paternal alloantigens by the embryo/placenta, we also depleted Aire+ cells in syngeneic pregnancies (groups #4 and #5). Resorption rates in Aire-depleted pregnancies tended to be higher than in controls (Fig. 1F), and importantly, the IUGR phenotype persisted with Aire+ cell depletion in syngeneic matings (Fig. 1G), indicating that Aire+ cells support tolerance to the conceptus, even when the fetus does not express paternal alloantigens. These results further suggested that the IUGR phenotype may be a more robust marker than resorption for Aire’s role in pregnancy complications.
We next investigated whether embryonic Aire+ cell depletion could account for these phenotypes, by breeding AireDTR males to WT females such that AireDTR expression (and potential depletion by DT) would be restricted to the embryo (group #6). Litter size (Fig. 1D), resorption rate (Fig. 1F) and average implantation site weight (Fig. 1G) were all similar to WT pregnancies without Aire+ cell depletion, indicating that the phenotypes of resorption and growth restriction are secondary to depletion of maternal, not embryonic, Aire+ cells.
Maternal immune response after Aire-expressing cell depletion
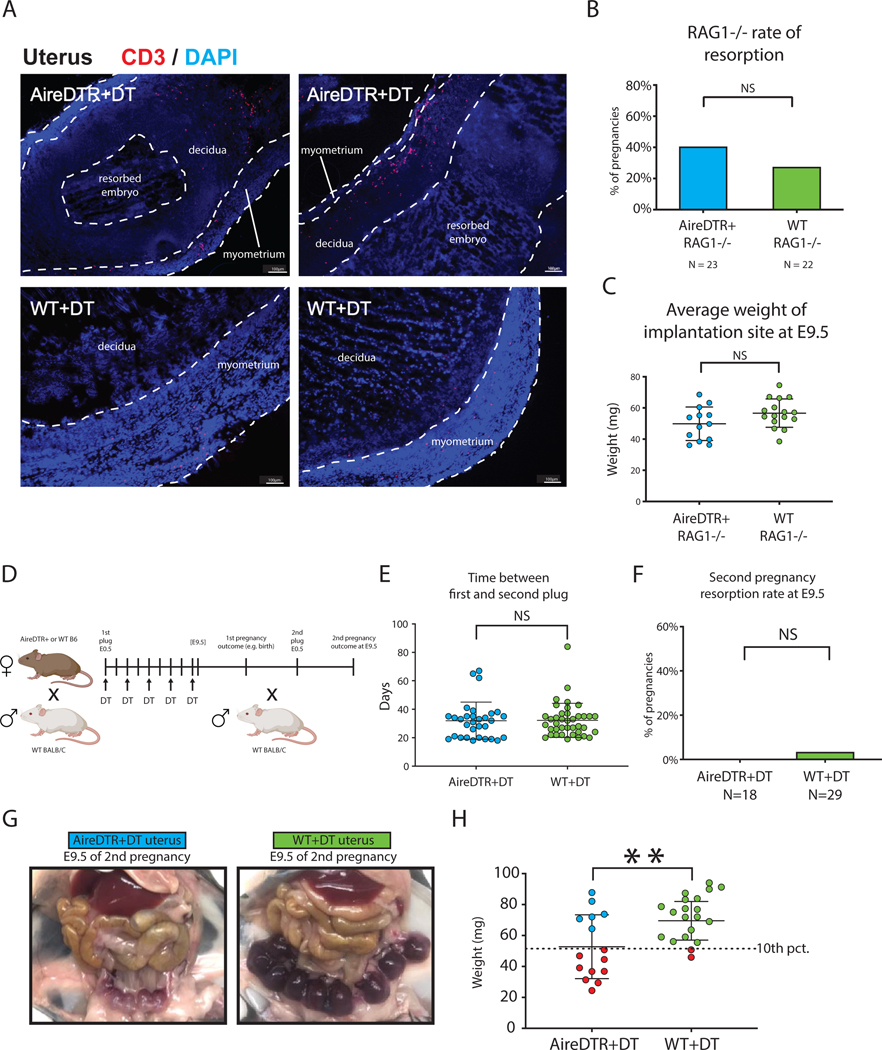
Immunofluorescent microscopy of the uterus in these experiments demonstrated marked infiltration of CD3+ cells into the myometrium (Fig 2A), suggesting an immunologic mechanism. To formally test whether the adaptive immune system was required for pregnancy complications in Aire-ablated mice, we next crossed the AireDTR transgene onto RAG1−/− mice, which lack functional T and B cells. AireDTR+RAG1−/− and AireDTR-RAG1−/− dams did not differ significantly in implantation rate, litter size, resorption rate or average implantation site weight at E9.5 (Fig. 2B,C, Fig. S1A, B). RAG1−/− dams were less likely to become pregnant after mating and had higher rates of baseline resorption, regardless of Aire+ cell ablation (Fig. S1A), which we have noted in prior experiments with this strain. These findings indicate a role for maternal adaptive immunity in the pregnancy complications observed in AireDTR+DT pregnancies.
Figure 2. The maternal adaptive immune system mediates pregnancy complications associated with Aire+ cell ablation.
(A) Immunofluorescence images showing infiltration of CD3 T cells in the uterus after Aire depletion. Scale bar = 100um. (B) Resorption rate and (C) average implantation site weight at E9.5 in AireDTR+RAG1−/− and AireDTR-RAG1−/− allogeneic pregnancies, treated with DT. (D) Schematic of experimental design: AireDTR and WT allogeneic pregnancies were treated with DT until E9.5, and the pregnancy was continued to term. Dams were bred a second time without DT treatment, and second pregnancy outcomes observed at E9.5. (E) Days between plug of first pregnancy and plug of second pregnancy. (F) Resorption rate of second pregnancy. (G) Representative gross pathology of the uterus at E9.5 of the second pregnancy. (H) Average implantation site weight at E9.5 of the second pregnancy. Dotted line indicates 10th percentile of implantation site weight in the WT+DT group. Red circles indicate concepti below the 10th percentile. ns = not statistically significant, p > 0.05, ** p < 0.01 by Fisher’s exact test in (B, F) and unpaired t-test in (C, E, H).
To further confirm whether this phenotype was immunologic in origin, we next asked whether there was a memory component, by examining the outcomes of second pregnancies, i.e. repeat exposure to PAAs, after selective Aire+ cell ablation only during the first pregnancy. AireDTR and WT females were treated with DT during their first pregnancy, bred a second time without DT exposure, and harvested at E9.5 (Fig. 2D). Time between first plug and second plug, implantation rate, litter size, and resorption rate did not differ significantly (Fig. 2E, F; Fig. S1C,D), but importantly, a severe IUGR phenotype occurred selectively in AireDTR+ dams, indicating a memory response to PAAs (Fig. 2G, H). These experiments further support the immunologic nature of the IUGR phenotype.
As patients with Aire mutations and AireKO mice develop systemic autoimmunity, especially in endocrine organs (21), we thought this pregnancy phenotype might be a secondary result of autoimmunity targeted against other endocrine organs. However we found no evidence of lymphocytic infiltration or other pathology in AireDTR+DT and WT+DT maternal adrenal glands, pancreatic islets, pituitary glands, or ovaries during the short course of these experiments (Fig. S2A). We also found no significant differences in the number of corpora lutea, which produce the progesterone required to maintain pregnancy (Fig. S2B). To investigate whether maternal progesterone deficiency made AireDTR+DT pregnancies vulnerable to resorption, we measured maternal serum progesterone at E6.5 and studied correlation with future pregnancy outcomes (live vs resorbed embryos) at E9.5. There were no significant differences between AireDTR+DT and WT+DT maternal serum progesterone at E6.5, regardless of E9.5 pregnancy outcome (Fig. S2C). To rule out a role for progesterone deficiency in this phenotype (22), we next gave exogenous progesterone to dams on days E4.5-E8.5 (peri-implantation) which did not prevent resorption or IUGR in AireDTR+DT dams (Fig. S2D,E). Thus, pregnancy complications in this model do not appear to be due to systemic autoimmunity or progesterone deficiency.
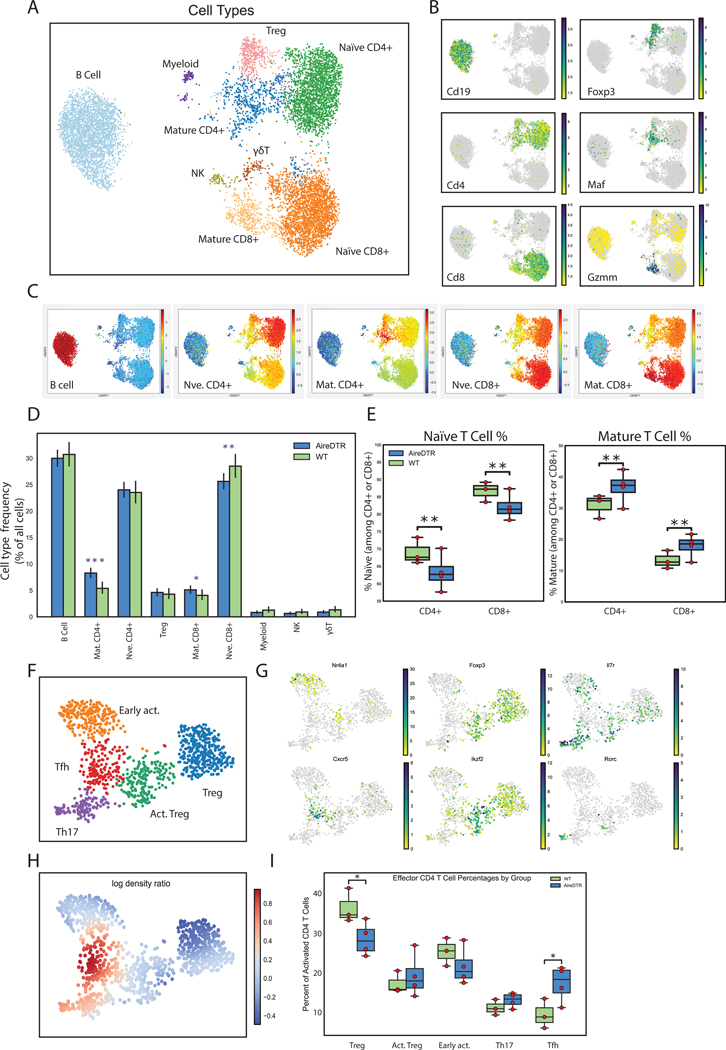
To better understand the immune mechanisms underlying this phenotype we next performed single-cell RNA sequencing (scRNAseq) on leukocytes obtained from the secondary lymphoid organs (uterine and non-uterine-draining lymph nodes) of WT and AireDTR pregnant dams during allogeneic pregnancy. All samples merged well (Fig. S3), and we were able to identify distinct lymphoid and myeloid populations (Fig. 3A). Feature mapping and per-cell similarity projections against reference populations from the ImmGen database (23) allowed us to clearly validate the identity of subpopulations of naïve, mature, and regulatory lymphoid and myeloid cells (Fig. 3B–D, Fig S4A,B). Strikingly, analysis of differential population distributions in WT and AireDTR mice demonstrated a significant shift from naïve to effector CD4+ and CD8+ T cells as a result of the loss of Aire-expressing cells (Fig. 3D, E). Among mature CD4+ T cells, there was a significant shift away from Tregs and toward mature T effectors (Fig. 3F–H), most notably T follicular helper (TFH) and Th17 cells (Fig. 3I). Together, these results are consistent with the idea that Aire+ cell ablation leads to loss of normal immune homeostasis, with increased activation of CD4+ and CD8+ T effectors and a concomitant shift from regulatory to effector lymphocyte populations in maternal lymph nodes.
Figure 3. Ablation of Aire+ cells leads to increased T-cell activation, and relative expansion of effector TFH and Th17 populations.
(A) Leiden clustering of pooled scRNAseq samples from DT-treated WT and AireDTR mouse brachial and uterine lymph nodes, populations identified as indicated. (B) Feature plots for indicated genes. (C) Per-cell projection of cosine similarity scores to indicated ImmGen reference cell types. (D) Cell type frequency (among all cells) in WT (green, n=3) and AireDTR (blue, n=4) mice. (E) Relative proportion of naïve vs. mature populations among CD4+ and CD8+ T cells; individual mice shown as yellow dots. (F) Leiden clustering of mature CD4+ and Treg populations from (A), above, with populations identified as indicated. (G) Feature plots for indicated genes. (H) Differential density plot showing distribution of increasing (red) or decreasing (blue) populations in AireDTR relative to WT (I) Box plots showing frequency per cell type among mature and regulatory populations shown in (F). Individual mice shown as yellow dots. * = p<0.05, ** = p<0.005 by unpaired t-test.
Differentiating the role of mTECs versus eTACs in maternal-fetal tolerance
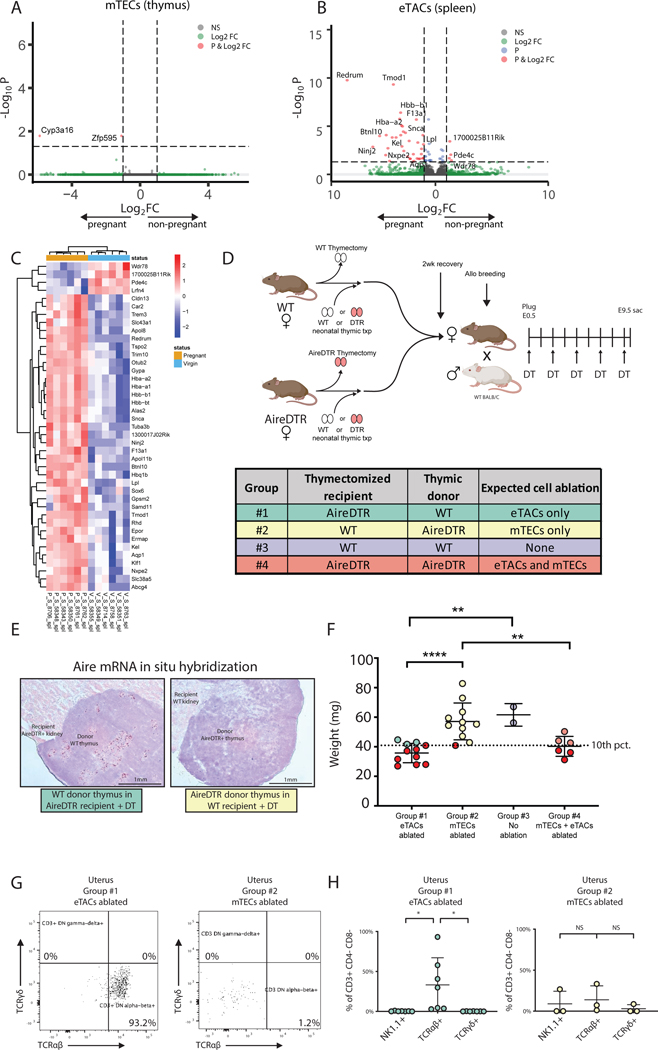
Despite the well-established significance of Aire in the thymus, the short duration of Aire+ cell ablation and the specific expansion of TFH populations in the previous experiment suggested a potential role for eTACs in this process; TFH cells depend on ICOS/ICOS-ligand signaling and mediate T cell-B cell crosstalk (24) and eTACs are known to cluster at T-B boundary regions and express high levels of ICOS-L (5). Further, loss of extrathymic Aire has previously been suggested to lead to expansion of TFH populations (25). We therefore investigated the relative contributions of thymic and extrathymic Aire-expressing cells to this pregnancy phenotype. We first investigated transcriptional changes in Aire+ cells in mTECs and eTACs using the Aire-driven Igrp-GFP (Adig) reporter mouse (5). We sorted mTECs and eTACs from virgin Adig females and E9.5 pregnant Adig dams for bulk RNA sequencing (Fig. S5A). While we found no differences in the frequency or absolute number of mTECs or eTACs (Fig. S5B), or of Aire expression in mTECs or eTACs (Fig. S5C) during pregnancy, we did find a significant number of differentially expressed (DE) transcripts during pregnancy in splenic eTACs, and almost no changes in mTECs (Fig. 4A–C, Fig. S5D–F). Pathway analysis of splenic DE genes revealed enrichment for erythrocyte development (Fig. S5G) and included a number of putative PAAs such as embryonic hemoglobin theta. While these transcriptional changes may represent eTAC-intrinsic expression of PAAs, pregnancy also causes increased maternal hematopoiesis (26) as well as passage of fetal nucleated RBCs into the maternal circulation (27) and the immunologic relevance of this phenomenon requires further study.
Figure 4. eTACs express PAAs during normal pregnancy and prevent growth restriction.
(A,B) Volcano plots and (C) heat map of differentially expressed genes (adj pval < 0.05, abs(log2FC) > 1), by bulk RNA sequencing of CD3- CD19- NK1.1- MHCIIhi GFP+ cells sorted from thymus (mTECs) or spleens (eTACs) of E9.5 pregnant (P, n=5) and virgin (V, n=6) Adig females. (D) Experimental design: AireDTR and WT females underwent thymectomy and received either a WT or AireDTR donor thymus transplant, and then were bred to allogeneic males and treated with DT every other day until E9.5. (E) In situ hybridization for Aire mRNA on transplanted WT thymus (left) and AireDTR+ thymus (right) after DT treatment. (F) Average implantation site weight at E9.5. Dotted line =10th percentile of implantation weight of mTEC-ablated group #2. Red circles indicate concepti below the 10th percentile. (G) Representative flow cytometry plots and (H) percentages of CD3 subsets in the uteri of eTAC or mTEC ablated dams. * p<0.05, ** p < 0.01, **** p < 0.0001 by Tukey’s multiple comparison test.
However, these results prompted us to ask formally whether the IUGR phenotype could be mapped to the ablation of mTECs or eTACs. We thus performed reciprocal thymic “swap” experiments in which we thymectomized WT and AireDTR females and simultaneously transplanted WT or AireDTR donor thymi under the kidney capsule. This approach allowed us to ablate mTECs, eTACs, both, or neither, in dams during pregnancy (Fig. 4D,E). We again found no significant differences in rates of implantation, litter size, or resorption among the groups (Fig. S6A,B). Surprisingly, the profound IUGR phenotype persisted, but only in eTAC-ablated, not mTEC-ablated, dams (Fig. 4F). Ablation of both cell types resulted in IUGR, whereas ablation of mTECs alone was equivalent to ablation of neither cell type, suggesting loss of eTACs was both necessary and sufficient for this phenotype. Interestingly, there was an increase in the proportion of αβTCR+ CD3+ CD4/8- cells, a previously described rare population with effector function (28, 29), in the uteri of dams with ablated eTACs (Fig. 4G,H). Thus, the phenotype of IUGR is functionally tied to loss of eTACs and associated with enrichment of adaptive immune cells in the uterus.
Discussion
These results suggest a previously unrecognized role for Aire expressing cells in maternal-fetal immune homeostasis. We have shown that depletion of Aire+ cells during pregnancy results in immune-mediated IUGR, that this phenomenon requires adaptive immunity, and has immunologic memory, and that a significant immune-activation phenotype is seen in the secondary lymphoid organs characterized by effector T cell, particularly TFH cell, activation. These results prompt the conclusion that the repertoire of antigens important for maternal-fetal tolerance must include syngeneic as well as paternal alloantigens expressed by the conceptus. Thus, these experiments provide a mechanistic insight into the known association of IUGR with autoimmune disease (1, 2).
Our findings also support a previously undescribed role for eTACs in maintaining maternal tolerance to the fetus. eTACs have previously been shown to mediate functional inactivation or deletion of cognate T cells and expansion of regulatory T cell populations (5, 6). In the context of pregnancy they may promote tolerance to the fetus through Aire-driven expression of PAAs, or by a range of means including suppression of T-cell activation and induction of regulatory populations. In the absence of eTACs, maternal effector T cells escape such regulation and infiltrate the uterus, contributing to adverse pregnancy outcomes (Fig. 5). Notably, IUGR has been linked to impaired maternal tolerance in early pregnancy, possibly as a result of impaired Treg induction (30).
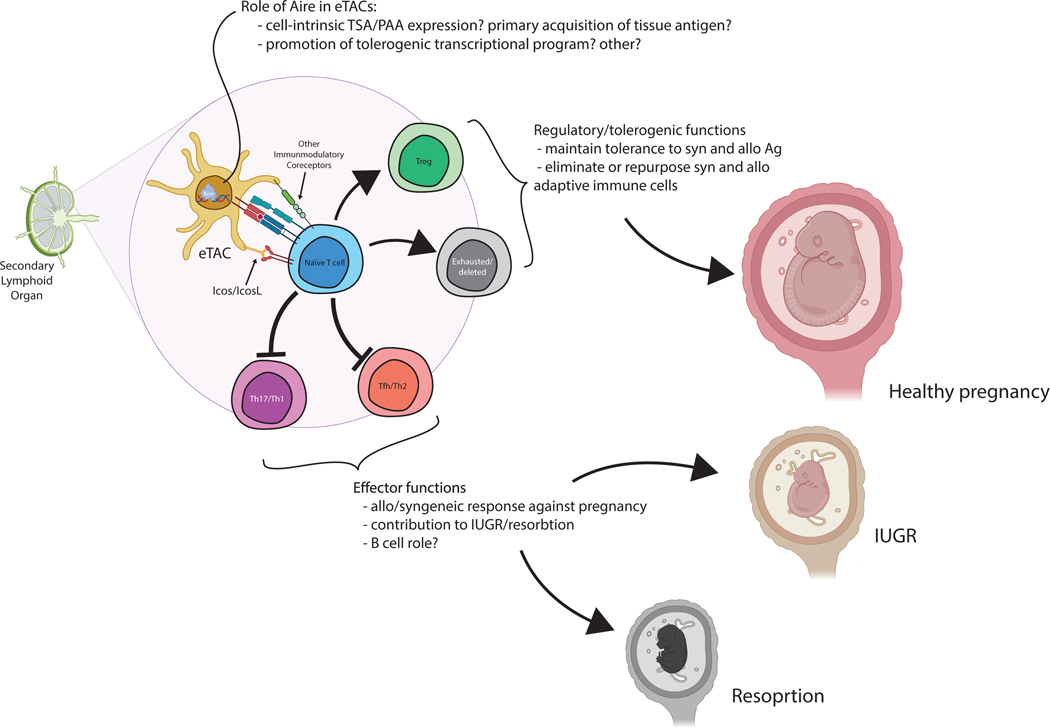
Figure 5. Proposed mechanism of the role of eTACs in maternal-fetal tolerance.
Extrathymic Aire-expressing cells may help maintain maternal-fetal tolerance through a range of mechanisms. Their loss leads to expansion of effector T cell populations, particularly TFH and Th17 cells, and results in immune-mediated IUGR and resorption. eTACs may interact with naïve T cells through a range of cell-surface receptors, and are known to express high levels of IcosL and other immunomodulatory genes. The precise role of Aire in this population remains to be determined, and may include expression of tissue-specific and pregnancy-associated antigens, induction of an immunoregulatory phenotype, or other unknown means. Both resorption and IUGR can result from breakdown of this immune homeostasis mechanism. This illustration was generated using biorender.com.
A critical aspect of maternal-fetal tolerance is tolerance to male-specific antigens (31). Some of these could be considered PAAs: encoded in the maternal genome but only expressed by male-specific tissues and thus only encountered during pregnancy with a male fetus. For example, autosomal prostate-specific antigens could be expressed in the male fetus but are potentially previously unseen antigens for the maternal immune system. Interestingly, however, Aire-dependent Tregs specific to prostate-specific antigen are found in both male and female mice, despite the lack of prostate tissue in females (32). Conversely, Y-chromosome-encoded minor antigens, such as H-Y, are also present in both allogeneic and syngeneic pregnancies, but these are not PAAs as the maternal genome does not include a Y-chromosome. In our AireDTR model, we observed IUGR or resorption of all implantation sites, as opposed to 50% of implantation sites if male embryos were selectively affected, as we have seen in a prior model of allospecific resorption (33). However, as expansion of antigen-specific effector T cells against a subset of the litter can have an adverse impact on the entire pregnancy in other settings (34, 35), the potential contribution of maternal immune reactivity against male antigens when Aire-expressing cells are depleted deserves further investigation.
The precise role of Aire in this process also remains unclear: while it may promote expression of PAAs, it is also possible that eTACs support pregnancy independent of the action of the Aire gene itself, or that Aire plays a different role in this population; further work is required to understand the cell-intrinsic role of Aire itself in peripheral tolerance induction.
However, the concept that Aire-expressing cells, traditionally recognized for their role in immune self-tolerance in mTECs, also play a role outside the thymus in reproductive fitness suggests a previously undescribed mechanism for maintaining maternal-fetal tolerance. More broadly, this demonstrates that loss of extrathymic Aire-expressing populations leads to a breakdown in normal immune homeostasis. If Aire-driven expression of PAAs in eTACs induces maternal tolerance to the embryo and placenta, this evolutionally important mechanism may be hijacked by cancers that express PAAs, and could explain why those cancers are so aggressive (36). Better understanding these systems may lead to therapies targeted to Aire-expressing populations to protect pregnancy and across a wide range of clinical applications.
Materials and Methods
Study Design
The aim of this study was to determine the role of maternal Aire-expressing cells during pregnancy. We used the AireDTR transgenic mouse and DT injections to ablate Aire-expressing cells during the first half of pregnancy, and confirmed mTEC and eTAC depletion using flow cytometry and in situ hybridization. To test if the pregnancy phenotype was due to progesterone deficiency, we gave pregnant AireDTR and WT mice supplemental progesterone. To test if the adaptive immune system was responsible for the observed phenotype, we bred AireDTR.RAGKO mice. We studied at least 20 plugged females in each experimental group to power the study given the rate of pregnancy loss in unmanipulated controls. To investigate the maternal immune response of Aire+ cell depletion, we used immunofluorescent microscopy, flow cytometry, and single-cell RNA sequencing to analyze T cells from the thymus, lymph nodes, spleen, and uterus of AireDTR+DT and WT+DT pregnant mice. To compare mTEC and eTAC transcripts from pregnant and nonpregnant mice, we used Adig transgenic mice and flow cytometry to isolate the mTECs and eTACs, and bulk RNA sequencing to compare the populations. Finally, to differentiate the effect of mTEC loss versus eTAC loss, we performed thymectomies and thymus transplants on AireDTR and WT mice before pregnancy.
Mice and Genotyping
All DNA primers were synthesized by Integrated DNA Technologies (Coralville, IA). C57BL/6 Aire-DTR transgenic mice were acquired from M. Anderson and the transgene was maintained in heterozygosity for all experiments. Offspring were screened for the presence of the AireDTR transgene using the genotyping primers 5’-GGACCTTTTGAGAGTCACTTTATCCT-3’ and 5’-CCCGTGCTCCTCCTTGCT-3’. AireDTR genotyping was also performed by Transnetyx using real-time PCR. C57BL/6 Adig transgenic mice were also acquired from M. Anderson and maintained in heterozygosity. Offspring were screened for the presence of the IGRP-GFP transgene using the genotyping primers 5’-AAGTTCATCTGCACCACC-3’ and 5’-TCCTTGAAGAAGATGGTGCG-3. AireDTR genotyping was also performed by Transnetyx using real-time PCR. C57BL/6 mice were obtained from Charles River Laboratories (Wilmington, MA). BALB/c, and RAG1−/− (B6.129S7-Rag1tm1Mom/J) mice were acquired from The Jackson Laboratory (Bar Harbor, ME; JAX stock#002216). RAG1−/− mice were crossed with Aire-DTR transgenic mice to achieve RAG1−/−Aire-DTR+/− experimental females, with RAG1−/−Aire-DTR−/− females as controls. RAG1 genotyping was performed by Transnetyx using real-time PCR. Males were mated with individual females once per week. Experimental females were bred to BALB/c males in allogeneic breedings or C57BL/6J males in syngeneic breedings, unless otherwise noted. The day of the copulation plug was designated E0.5. All mice were maintained in microisolator cages and treated in accordance with NIH and American Association of Laboratory Animal Care standards, and consistent with the animal care and use regulations of the University of California, San Francisco.
Diphtheria Toxin Treatment
Diphtheria toxin (Enzo Life Sciences, Farmingdale, NY) was administered intraperitoneally at a dose of 15–25 ng/g in sterile PBS, every other day, from E0.5 until E8.5.
Lymphocyte Preparation for Flow Cytometry and RNA Sequencing
Lymphocytes were prepared for flow cytometry by mashing thymi, lymph nodes, and spleens, which were then filtered through a 70-micron cell-strainer. Red blood cells were lysed by incubating peripheral blood and spleen samples in ACK lysing buffer (Lonza BioWhittaker, Basel, Switzerland) for 10 minutes. If pregnant, embryos were removed from the maternal uterus, which was then minced with razor blades and digested for 15 minutes at 37°C with 5% CO2 in 2 mg/mL collagenase D (Roche, Basel, Switzerland) and 10 μg/mL DNase I recombinant (Roche, Basel, Switzerland) as previously described (33). Digested cells were resuspended in cold FACS buffer (DPBS containing 0.5% BSA and 0.2 mM EDTA) and filtered through a 70-micron cell-strainer. Cells were resuspended in anti-Fc receptor blocking antibody clone 2.4G2 for 10 minutes, then incubated on ice for 30 minutes with antibody cocktails. Samples were fixed, permeabilized, and incubated with 1:100 FoxP3 antibody using an intranuclear staining kit (Thermo Fisher, Waltham, MA). Stained samples were acquired on an LSRII (BD Biosciences, San Jose, CA), and flow cytometry data were analyzed using FlowJo (FlowJo LLC, Ashland, OR) and Prism software (GraphPad Software, San Diego, CA).
For bulk RNA sequencing, stromal cells were prepared for cell sorting by mincing thymus, lymph nodes, and spleen with razor blades and then digesting for 30 minutes at 37°C in 100ug/mL DNase I (Roche) and Liberase TM (Roche) before filtering through a 70-micron cell-strainer. Density-gradient centrifugation was used to enrich for stromal cells, using a three-layer Percoll PLUS gradient (Sigma) with specific gravities of 1.115, 1.065, and 1.0. Cells isolated from between the 1.065 and 1.0 layer were resuspended in anti-Fc receptor blocking antibody clone 2.4G2 for 10 minutes, then incubated on ice for 30 minutes with antibody cocktail. 4’,6-diamidino-2-phenylindole (DAPI) was added to exclude dead cells before sorting on a FACSAria II (BD Biosciences, San Jose, CA ). CD3- CD19- NK1.1- MHCII+ GFP+ cells were sorted directly into cold RPMI with 2% FBS, washed with PBS twice, and then resuspended in lysis buffer and snap frozen for RNA isolation and sequencing.
For single cell RNA sequencing, lymphocytes were prepared by mashing lymph nodes, which were then filtered through a 70-micron cell-strainer. The maternal uterus was minced with razor blades and digested for 15 minutes at 37°C with 5% CO2 in 2mg/mL collagenase D (Roche, Basel, Switzerland) and 10ug/mL DNase I recombinant (Roche, Basel, Switzerland) as previously described (33). Digested cells were resuspended in cold FACS buffer (DPBS containing 0.5% BSA and 0.2 mM EDTA) and filtered through a 70-micron cell-strainer. Density-gradient centrifugation was used to enrich for lymphocyte cells, using a three-layer Percoll PLUS gradient (Sigma) with specific gravities of 1.115, 1.065, and 1.0. Cells isolated from between the 1.065 and 1.115 layer were resuspended in anti-Fc receptor blocking antibody clone 2.4G2 for 10 minutes, then incubated on ice for 30 minutes with antibody cocktail. Live, CD45+ CD3+ cells were sorted into PBS with 2% BSA following the 10x Genomics protocol for multiplexing and single cell sequencing. Libraries were constructed using the Single Cell 3′ Library Kit V3 (10x Genomics). Once prepared, indexed complementary DNA (cDNA) libraries were sequenced with paired-end reads on an Illumina NovaSeq 6000 (Illumina).
Single cell sequencing analysis:
Pre-processing and Demultiplexing
Gene expression matrices were generated by pre-processing the raw reads from each batch with the kallisto|bustools pipeline (37–39). Tag count matrices were generated from the raw reads of the barcoding experiment with kITE (40) and kallisto | bustools. To assign tag identity to a cell, we calculated the proportion of counts from each tag in the cell. A multinomial confidence interval was calculated for each proportion using the Goodman approximation method implemented with Python package statsmodels.stats.proportions. A tag was assigned to a cell if the lower confidence interval value of the most abundant tag was two times the high confidence interval value of the next most abundant tag. This strict measure for cell identity allows us to filter out doublets. Additionally, cell barcodes a) with more than 5% mitochondrial reads, or b) with a total number of counts or number of genes above the 99.5 percentile or c) with less than 10 observed counts were removed from each batch. Uterus samples were further excluded from the dataset due to a low number of detected cells (only 20 cells were confidently demultiplexed and assigned to uterus samples across batches, compared to 6919 and 4442 cells assigned to brLN and udLN respectively). The resulting pre-processed and demultiplexed count matrices from each experiment were concatenated for further analysis and downstream processing with scanpy (41).
Normalization, Clustering and Visualization
The demultiplexed data were normalized to have 10,000 counts/cell and were log1p-transformed. Highly variable genes were calculated using the scanpy function highly_variable_genes using Seurat (42) flavor with the default parameters (min_mean=0.0125, max_mean=3 and min_disp=0.5). Only highly variable genes were used for further analysis. The total number of counts per cell was regressed out and the gene expression matrix was scaled using scanpy function scale with max_value =10. Dimensionality reduction was performed using principal component analysis with 50 principal components. Batch balanced kNN (43), implemented with scanpy’s function bbknn, was used to compute each cell’s top neighbors and normalize batch effects. The batch corrected cells were clustered using the Leiden algorithm (44) and projected in to two dimensions with UMAP (45) for visualization. Initial cluster identity was determined by finding marker genes with differential expression analysis preformed using a t-test on log1p-transformed raw counts with scanpy function rank_genes_groups. Clusters associated with a) erythrocyte markers b) co-expression of B-cell and T-cell markers and clusters of ambiguous identity representing less than 0.1% of the total number of cells were filtered out.
Cell type annotation / ImmGen similarity scores
In order to assign identities to the resulting cell subpopulations/clusters in an unbiased way, we developed an approach to query the similarity of single-cell RNA-seq profiles against all 224 publicly available reference microarray profiles in the Immunological Genome Project (ImmGen) Database; all ImmGen primary data were downloaded from haemosphere (https://www.haemosphere.org/datasets/show). The similarity measure we used is based on the cosine distance between appropriately normalized microarray/gene expression profiles -considering only genes that were identified as highly variable in scRNAseq. For each reference subpopulation, the corresponding ImmGen microarray profile was normalized to unit sum. The single cell gene expression profiles were first scaled to 1e4 total counts and logarithmized before being normalized to unit sum. For each single cell profile, the resulting cosine similarities to all ImmGen profiles were further standardized (by removing the mean and scaling to unit variance), to obtain the final the final ImmGen similarity scores shown in Fig. 3B–D; Fig. S4A, B. Final cell type annotation was carried out in a semi-supervised way, cross-validating the resulting ImmGen scores with subpopulation-specific gene expression of known markers (Fig. 3F, G; Fig. S4C).
Cell-type proportions
We modelled treatment (AireDTR vs WT) as an explanatory variable in a binomial generalized linear model (binomial GLM) in which a count from the cell type of interest was a successful outcome in a binomial experiment. We fit the binomial GLM with Python package statsmodels.stats to count data from each biological replicate. Cell-type composition changes were considered to be significant if the p-value from the binomial GLM was less than 0.05. Multinomial confidence intervals for each cluster proportion were calculated via the Goodman method.
UMAP Differential Density Estimation
To visualize the differences in density across cell sub-populations between AireDTR and WT samples, we used an approach based on Kernel Density Estimation (KDE). The AireDTR and WT densities were computed independently using the UMAP coordinates of the corresponding cells as input and were extrapolated to the entire 2-dimensional space, using the nonparametric kernel_density.KDEMultivariate() function from the statsmodels Python package. For each point in the UMAP space we then calculated and visualized the logarithm of the estimated densities ratio, log(AireDTR_density/WT_density), as a proxy for the likelihood of a UMAP region being associated with increased (positive log density ratio) or decreased (negative log density ratio) proportions of AireDTR cells.
P-value significance levels for plots:
***: p<=0.0005, **: 0.0005<p<=0.005, *: 0.005 <=p< 0.05
Bulk RNA sequencing and Analysis
We employed standard mRNA-seq protocols used by the UCSF Functional Genomics sequencing core. Following library construction with Smart-Seq/NexteraXT (Illumina, San Diego, CA) cDNA fragments underwent 50-basepair single-end sequencing on an Illumina HiSeq 4000. Sequencing provided was 2.53 billion total reads with an average of 83.6% of these reads aligning uniquely to the human genome (Ensembl Human GRCh38.78). STAR (46) version 2.5.2b was used to align reads to the mouse genome, version GRCm38.78, and no adapter clipping or filtering was performed before alignment. Reads uniquely mapped to known mRNAs were used to assess expression changes between genes. Pairwise comparisons were made between pregnant vs virgin mice in the thymus, spleen, uterine-draining lymph node, and non-uterine-draining lymph node, in using DESeq2 (47) in R. Transcripts were combined by geneID and transcripts with 0 counts were removed resulting in 21,478 transcripts across 48 samples. FDR multiple hypothesis correction was applied and genes with adjusted p-value < 0.05 and absolute value log Fold Change > 1 were considered significant. We used 6 mice per biological replicate. Pathway analysis was carried out with ClusterProfiler (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339379/) using the KEGG database (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5210567/).
Histology
For hematoxylin and eosin staining, tissues were fixed overnight at 4°C in 10% neutral buffered formalin (Thermo Fisher, Waltham, MA) and then dehydrated in 70% ethanol before being sent to HistoWiz (Brooklyn, NY) for paraffin embedding, 5 μm sectioning, staining, and evaluation for lymphocyte infiltration by a pathologist employed by HistoWiz who was blinded to experimental groups. For in situ hybridization, fresh frozen tissues were sectioned at 10 μm and then fixed overnight at 4°C in 10% neutral buffered formalin before proceeding with RNAscope RED 2.5 HD Chromogenic Assay kit (Advanced Cell Diagnostics, Newark, CA) for detection of Aire mRNA. For immunofluorescence, fresh frozen tissues were sectioned at 10 μm and fixed in 1:1 methanol:acetone solution for 20 min at −20°C, blocked with goat serum, and then incubated in Rabbit anti-CD3ε Clone SP7 (Invitrogen) overnight. Secondary staining was completed the next day using the goat anti-rabbit IgG AF555 Tyramide SuperBoost Kit (Invitrogen), and mounted with Fluoromount-G mounting medium with DAPI (Invitrogen).
Serum progesterone level measurements:
Serum was collected by submandibular bleeds and stored at −80°C before being sent to Cayman Chemical Company for enzyme-linked immunoassay (Ann Arbor, MI).
Progesterone supplementation during pregnancy:
Progesterone (Sigma-Aldrich, St, Louis, MO) was dissolved in sesame seed oil (Sigma-Aldrich, St. Louis, MO) and administered subcutaneously at a dose of 2 mg/100 μL, daily, from E4.5 until E8.5, as described in (22).
Thymectomy and Thymic Transplant
Donor thymi were collected from euthanized neonates and then cultured with 1.35 mM 2′-deoxyguanosine monohydrate (Sigma) for 5–7 days to deplete endogenous lymphocytes. 4–6- week-old recipient mice were anesthetized and a small incision was placed along the midline of the first three ribs to enter the thorax and expose the area around the thymus. A sterile pipette was inserted into the thorax and the thymus was removed by vacuum. The incision was closed with surgical clips. A left flank incision was used to expose the left kidney, and the donor neonatal thymus was transplanted under the kidney capsule. The peritoneum was closed with sutures and the skin with surgical clips, which were removed 14 days post-procedure, at which time mice were bred for experiments. At time of sacrifice, the thorax was examined to confirm complete thymic excision and the kidney was examined for presence of the transplanted thymus; if the transplanted thymus was not present, that mouse was excluded from experimental analysis.
Statistical Analysis
Implantation rate (percentage of plugged females with visible implantation sites at E9.5) and resorption rate (percentage of completely resorbed pregnancies of all pregnancies with visible implantation sites at E9.5) were analyzed using Fisher’s exact test. Litter size, weight of implantation site, and percentages of flow cytometry cell subsets were analyzed using by Tukey’s multiple comparison test (when comparing three or more experimental groups) or unpaired t-test (when comparing two experimental groups). For all tests, not significant = p > 0.05 and significant = * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Error bars represent normal-based 95% confidence interval. For single cell RNA sequencing, we fit the binomial GLM with Python package statsmodels.stats to count data from each biological replicate. Cell-type composition changes were considered to be significant if the p-value from the binomial GLM was less than 0.05. Multinomial confidence intervals for each cluster proportion were calculated via the Goodman method. For bulk RNA sequencing, FDR multiple hypothesis correction was applied and genes with adjusted p-value < 0.05 and absolute value log Fold Change > 1 were considered significant. We used 6 mice per biological replicate.
Supplementary Material
Raw data file (Excel spreadsheet)
Figure S1. Implantation rates and litter sizes are not affected by Aire+ cell ablation in RAG1−/− dams, or in B6.AireDTR dams’ second pregnancy after Aire+ cells were ablated during their first pregnancy
Figure S2. Aire+ cell ablation does not lead to systemic maternal endocrinopathy, and pregnancy complications are not secondary to progesterone deficiency
Figure S3. Batching and clustering of samples used in scRNAseq analysis.
Figure S4. scRNAseq cluster identities defined by transcriptional phenotype
Figure S5. eTACs, not mTECs, differentially express PAAs during pregnancy
Figure S6. No difference in implantation rate or litter size in thymic transplant recipients with mTEC and/or eTAC ablation
Acknowledgments:
We thank members of the MacKenzie, Gardner, and Anderson labs and Drs. Adrian Erlebacher and Susan Fisher for helpful discussions. We thank Maria Clark, Carlo Eikani, Jasmine Valencia and Bowen Wang for technical assistance.
Funding:
Supported by NIH R01 A125452 and R01 AI145858 to T.C.M., and a Howard Hughes Medical Research Fellows Program Grant to E.G.B., and the UCSF Sandler PSSP and ASTS Fellowship in Transplantation grants to J.M.G.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The sequencing data for this study have been deposited in the database GEO and can be accessed using accession numbers GSE174521 (scRNAseq) and GSE174730 (bulk RNAseq. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes:
- 1.Lin HC, Chen SF, Lin HC, Chen YH, Increased risk of adverse pregnancy outcomes in women with rheumatoid arthritis: a nationwide population-based study. Ann Rheum Dis 69, 715–717 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Sen A, Kushnir VA, Barad DH, Gleicher N, Endocrine autoimmune diseases and female infertility. Nat Rev Endocrinol 10, 37–50 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D, Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Malchow S, Leventhal DS, Lee V, Nishi S, Socci ND, Savage PA, Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity 44, 1102–1113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner JM, DeVoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, Johannes KP, Su MA, Chang HY, Krummel MF, Anderson MS, Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner JM, Metzger TC, McMahon EJ, Au-Yeung BB, Krawisz AK, Lu W, Price JD, Johannes KP, Satpathy AT, Murphy KM, Tarbell KV, Weiss A, Anderson MS, Extrathymic Aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4(+) T cells. Immunity 39, 560–572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyewski B, Derbinski J, Self-representation in the thymus: an extended view. Nat Rev Immunol 4, 688–698 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Dragin N, Bismuth J, Cizeron-Clairac G, Biferi MG, Berthault C, Serraf A, Nottin R, Klatzmann D, Cumano A, Barkats M, Le Panse R, Berrih-Aknin S, Estrogen-mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Invest 126, 1525–1537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu ML, Bakhru P, Conley B, Nelson JS, Free M, Martin A, Starmer J, Wilson EM, Su MA, Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun 7, 11350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soumya V, Padmanabhan RA, Titus S, Laloraya M, Murine uterine decidualization is a novel function of autoimmune regulator-beyond immune tolerance. Am J Reprod Immunol 76, 224–234 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Warren BD, Ahn SH, McGinnis LK, Grzesiak G, Su RW, Fazleabas AT, Christenson LK, Petroff BK, Petroff MG, Autoimmune Regulator (AIRE) is required in female mice for optimal embryonic development and implantation. Biol Reprod 100, 1492–1504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou X, Xhang Y, Wang X, Zhang R, Yang W, The role of AIRE deficiency in infertility and its potential pathogenesis. Front Immunol 12, 641164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barinova IV, Burumkulova FF, Shidlovskaia NV, Bashakin NF, Petrukhin VA, Kondrikov NI, [Placental alterations in pregnant women with autoimmune polyglandular endocrinopathy]. Arkh Patol 75, 14–17 (2013). [PubMed] [Google Scholar]
- 14.Logutova LS, Petrukin VA, Burumkulova FF, Shidlovskaya NV, Autoimmune polyglandular syndrome type 1 and pregnancy: Clinical types. Russian Bulletin of Obstetrics and Gynecology 3, 59–64 (2015). [Google Scholar]
- 15.Metzger TC, Khan IS, Gardner JM, L Mouchess M, Johannes KP, Krawisz AK, Skrzypczynska KM, Anderson MS, Lineage tracing and cell ablation identify a post-Aire-expressing thymic epithelial cell population. Cell Rep 5, 166–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jasti S, Warren BD, McGinnis LK, Kinsey WH, Petroff BK, Petroff MG, The autoimmune regulator prevents premature reproductive senescence in female mice. Biol Reprod 86, 110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paolino M, Koglgruber R, Cronin SJF, Uribesalgo I, Rauscher E, Harreiter J, Schuster M, Bancher-Todesca D, Pranjic B, Novatchkova M, Fededa JP, White AJ, Sigl V, Dekan S, Penz T, Bock C, Kenner L, Holländer GA, Anderson G, Kautzky-Willer A, Penninger JM, RANK links thymic regulatory T cells to fetal loss and gestational diabetes in pregnancy. Nature 589, 442–447 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen T, Darrasse-Jèze G, Bergot A, Courau T, Churlaud G, Valdivia K, Strominger JL, Ruocco MG, Chaouat G, Klatzmann D, Self-specific memory regulatory T cells protect embryos at implantation in mice. J Immunol 191, 2273–2281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shima T, Sasaki Y, Itoh M, Nakashima A, Ishii N, Sugamura K, Saito S, Regulatory T cells are necessary for implantation and maintenance of early pregnancy but not late pregnancy in allogeneic mice. J Reproductive Immunology 85 (2010): 121–129. [DOI] [PubMed] [Google Scholar]
- 20.Figueras F, Gardosi J, Intrauterine growth restriction: new concepts in antenatal surveillance, diagnosis, and management. Am J Obstet Gynecol 204, 288–300 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Husebye ES, Anderson MS, Kampe O, Autoimmune Polyendocrine Syndromes. N Engl J Med 378, 2543–2544 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Erlebacher A, Zhang D, Parlow AF, Glimcher LH, Ovarian insufficiency and early pregnancy loss induced by activation of the innate immune system. J Clin Invest 114, 39–48 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heng TS, Painter MW, Immunological Genome Project C, The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Bossaller L, Burger J, Draeger R, Grimbacher B, Knoth R, Plebani A, Durandy A, Baumann U, Schlesier M, Welcher AA, Peter HH, Warnatz K, ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J Immunol 177, 4927–4932 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Lindmark E, Chen Y, Georgoudaki A, Dudziak D, Lindh E, Adams WC, Loré K, Winqvist O, Chambers BJ, Karlsson MCI, AIRE expressing marginal zone dendritic cells balances adaptive immunity and T-follicular helper cell recruitment. J Autoimmun 42, 62–70 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Nakada D, Oguro H, Levi BP, Ryan N, Kitano A, Saitoh Y, Takeichi M, Wendt GR, Morrison SJ, Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchi DW, Zickwolf GK, Yih MC, Flint AF, Geifman OH, Erikson MS, Williams JM, Erythroid-specific antibodies enhance detection of fetal nucleated erythrocytes in maternal blood. Prenat Diagn 13, 293–300 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Cowley SC, Hamilton E, Frelinger JA, Su J, Forman J, Elkins KL, CD4-CD8- T cells control intracellular bacterial infections both in vitro and in vivo. J Exp Med 202, 309–319 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson M, Lycke N, A unique population of extrathymically derived αβTCR+ CD4− CD8− T cells with regulatory functions dominates the mouse female genital tract. J Immunology 170, 1659–1666 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Robertson SA, Care AS, Moldenhauer LM, Regulatory T cells in embryo implantation and the immune response to pregnancy. J Clin Invest 128, 4224–4235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn DA, Baltimore D, Pregnancy induces a fetal antigen-specific maternal T regulatory cell response that contributes to tolerance. PNAS, 107, 9299–9304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, Amit AS, Kang C, Geddes JE, Allison JP, Socci ND, Savage PA, Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wegorzewska M, Nijagal A, Wong CM, Le T, Lescano K, Tang Q, MacKenzie TC Fetal intervention increases maternal T cell awareness of the foreign conceptus and can lead to immune-mediated fetal demise. J Immunol 192, 1938–1945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moldenhauer LM, Diener KR, Hayball JD, Robertson SA, An immunogenic phenotype in paternal antigen‐specific CD8+ T cells at embryo implantation elicits later fetal loss in mice. Immunology and Cell Biology 95, 705–715 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Rowe JH, Ertelt JM, Xin L, Way SS, Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature 490, 102–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtan SG, Creedon DJ, Haluska P, Markovic SN. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clinic Proceedings 84, 985–1000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Melsted P, Ntranos V, Pachter L, The barcode UMI, set format and BUStools. Bioinformatics 35, 4472–4473 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Melsted P, Booeshaghi S, Gao F, Beltrame E, Lu K, Hjorleifsson KE, Gehring J, Pachter L, Modular and efficient pre-processing of single cell RNA-seq. BioRxiv 10.1101/673285, (2019). [DOI]
- 40.Gehring J, Hwee Park J, Chen S, Thomson M, Pachter L, Highly multiplexed single- cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nature biotechnology 38, 35–38 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, Park J-E, BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traag VA, Waltman L, van Eck NJ, From Louvain to Leiden: guaranteeing well- connected communities. Sci Rep 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW, Dimensionality reduction for visualizing single-cell data using UMAP. Nature biotechnology 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data file (Excel spreadsheet)
Figure S1. Implantation rates and litter sizes are not affected by Aire+ cell ablation in RAG1−/− dams, or in B6.AireDTR dams’ second pregnancy after Aire+ cells were ablated during their first pregnancy
Figure S2. Aire+ cell ablation does not lead to systemic maternal endocrinopathy, and pregnancy complications are not secondary to progesterone deficiency
Figure S3. Batching and clustering of samples used in scRNAseq analysis.
Figure S4. scRNAseq cluster identities defined by transcriptional phenotype
Figure S5. eTACs, not mTECs, differentially express PAAs during pregnancy
Figure S6. No difference in implantation rate or litter size in thymic transplant recipients with mTEC and/or eTAC ablation