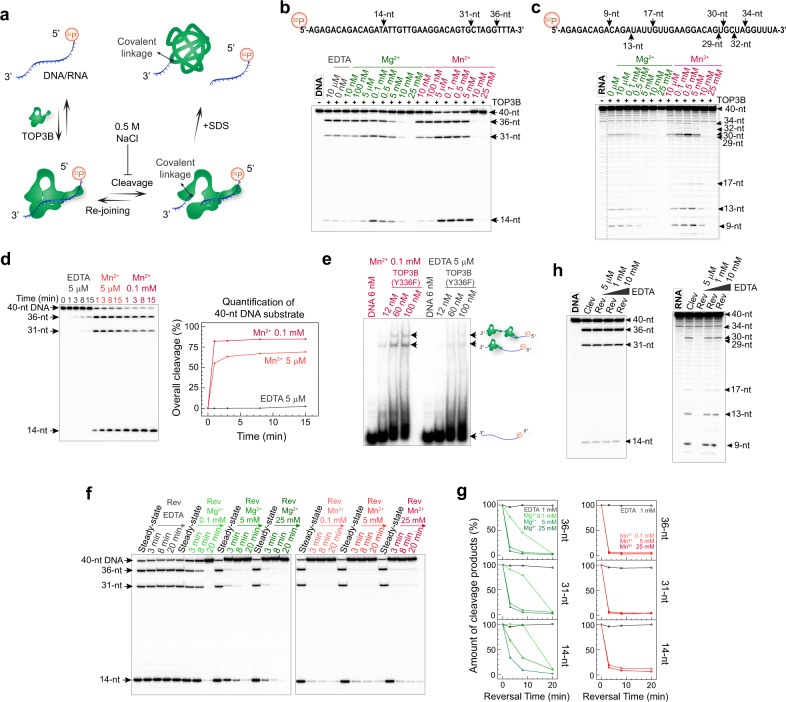
Fig. 1. TOP3B-mediated DNA and RNA cleavage and rejoining are differentially affected by Mg2+ and Mn2+.
a Schematic diagram showing DNA/RNA substrate labeling and TOP3B cleavage assay. b, c DNA and RNA cleavage by TOP3B at various Mg2+ and Mn2+ concentrations. Arrows indicate major DNA/RNA cleavage products and their corresponding cleavage sites in the substrate sequences. Gray dotted vertical line in c indicates combination of different lanes on the same gel with the same exposure condition. DNA cleavage sites were determined with sequence-specific DNA oligos as markers. RNA cleavage sites were mapped with RNA alkaline ladder and RNase T1 ladder (Suppl Fig. 2). d Time-course showing pre-steady-state DNA product formation in the absence of divalent cation (and with 5 μM EDTA) or with 5 μM or 0.1 mM Mn2+. Plot of DNA cleavage product versus time was obtained by analyzing the depletion of the 40-nt DNA substrate band. e EMSA assay with catalytic inactive mutant TOP3B-Y336F showing its DNA binding in the absence and presence of Mn2+. TOP3B retardation bands are indicated by arrows and cartoons. f Time-course of DNA end-rejoining by TOP3B at various Mg2+ and Mn2+ concentrations. Steady state levels of cleavage were obtained by pre-incubating TOP3B with DNA for 30 min in solutions containing 0.1 mM Mg2+ or Mn2+. Subsequent reversal steps (Rev) were initiated by addition of 0.5 M NaCl and indicated amounts of divalent ions or EDTA. g Plots show reversal of DNA cleavage products over time (percentage) obtained by quantification of individual DNA products in panel f. h DNA and RNA cleavage-reversal assays at various EDTA concentrations. Cleavage (Clev) products were generated by incubating TOP3B with DNA/RNA substrate for 30 min in the presence of 5 μM EDTA, followed by reversal analysis (Rev) by adding 0.5 M NaCl and the indicated amounts of EDTA for 5 min. All the results of the gel-based assays in (b–h) are representative results obtained from 2–4 independent experimental repeats.