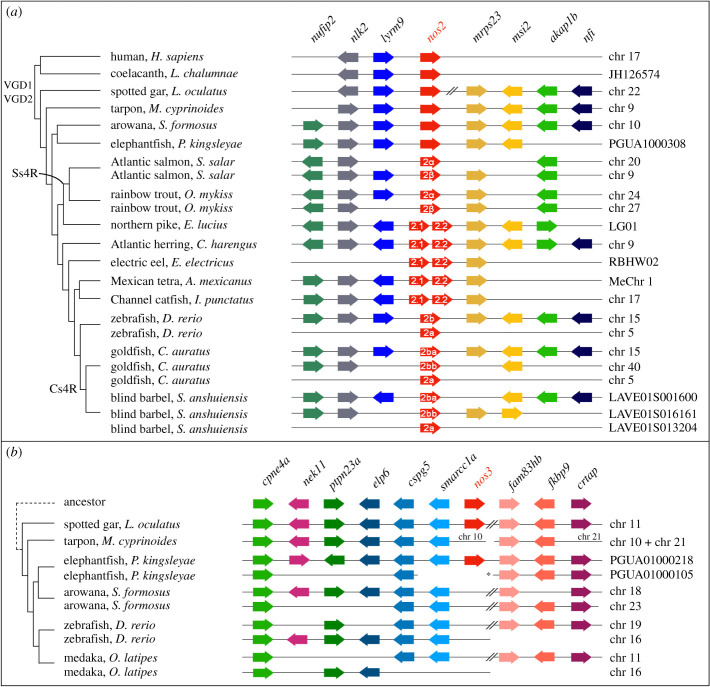
Figure 2.
Conserved microsynteny of nos2 and nos3. (a) The nos2 paralogues derived from different duplication modalities: carp-specific genome duplication (Cs4R) (nos2ba and nos2bb in the goldfish and blind barbel); salmonid-specific genome duplication (Ss4R) (nos2α and nos2β in the Atlantic salmon and rainbow trout); tandem gene duplication occurred independently in five lineages (nos2.1 and nos2.2 in the northern pike, Atlantic herring, electric eel, Mexican tetra and channel catfish). An additional nos2 duplicate (nos2a) is present in cyprinids (zebrafish, goldfish, and blind barbel) (see the electronic supplementary material, figure S2). (b) A conserved synteny map of genomic regions around the nos3 gene locus highlights the loss in Clupeocephala (including zebrafish and medaka), and in Osteoglossomorpha (arowana). Consecutive genes are represented as arrows and are colour coded according to their orthology and ohnology. The direction of arrows indicates gene transcription orientation. // indicates long-distance on the chromosome (>600 kb), * indicates scaffold 72 of the freshwater elephantfish genome [15]. (Online version in colour.)