Abstract
Comparative analyses of bats indicate that hibernation is associated with increased longevity among species. However, it is not yet known if hibernation affects biological ageing of individuals. Here, we use DNA methylation (DNAm) as an epigenetic biomarker of ageing to determine the effect of hibernation on the big brown bat, Eptesicus fuscus. First, we compare epigenetic age, as predicted by a multi-species epigenetic clock, between hibernating and non-hibernating animals and find that hibernation is associated with epigenetic age. Second, we identify genomic sites that exhibit hibernation-associated change in DNAm, independent of age, by comparing samples taken from the same individual in hibernating and active seasons. This paired comparison identified over 3000 differentially methylated positions (DMPs) in the genome. Genome-wide association comparisons to tissue-specific functional elements reveals that DMPs with elevated DNAm during winter occur at sites enriched for quiescent chromatin states, whereas DMPs with reduced DNAm during winter occur at sites enriched for transcription enhancers. Furthermore, genes nearest DMPs are involved in regulation of metabolic processes and innate immunity. Finally, significant overlap exists between genes nearest hibernation DMPs and genes nearest previously identified longevity DMPs. Taken together, these results are consistent with hibernation influencing ageing and longevity in bats.
Keywords: DNA methylation, epigenetic clock, Eptesicus fuscus, metabolism, immunity, longevity
1. Introduction
A variety of mammals save energy during periods of food shortage by entering into torpor, a physiological state characterized by reduced body temperature and metabolic rate that lasts a few hours or days [1,2]. More prolonged bouts of torpor that occur during the winter are referred to as hibernation, and occur in some rodents, bats, carnivores, hedgehogs, marsupials, tenrecs and primates [3–5]. Comparisons of gene expression in tissues taken from hibernating and non-hibernating animals have revealed that hibernation not only suppresses metabolic function [6–9] but also protects skeletal muscle from atrophy [10,11] and inhibits immune function [10,11]. Despite the widespread occurrence of hibernation among mammals, the genetic mechanisms regulating this process remain poorly understood in most species [12,13].
Recently, DNA methylation (DNAm) has been used to gain insight into patterns of gene regulation during hibernation [14,15]. DNA is methylated when a methyl group is added to a cytosine at cytosine–guanine dinucleotide (CpG) sites [16]. While this change can be functionally silent, DNAm can suppress transcription by recruiting proteins involved in gene repression [15,17] or by altering chromatin states to inhibit transcription factor binding [18]. Conversely, loss of DNAm, especially in promoter regions, can permit recruitment of proteins involved in gene activation and result in transcription. Examining patterns of DNAm in a tissue can, therefore, reveal repressed or elevated transcription of specific genes or gene pathways in response to environmental changes [16,19].
To date, DNAm has only been examined in a few hibernating rodent species. Compared to euthermic controls, global DNAm increased nearly twofold in brown adipose tissue [15] but decreased in liver and skeletal muscle in 13-lined ground squirrels, Ictidomys tridecemlineatus [14]. The presence of DNAm has also been used to assess tissue-specific transcription in a hibernation candidate gene (HP-27) in chipmunks (Tamias asiaticus); DNAm at an upstream stimulatory factor (USF)-binding site, which partially regulates transcription of HP-27, was reduced in the liver but increased in the kidneys and heart [20], which is consistent with liver-specific transcription for HP-27. In yellow-bellied marmots (Marmota flaviventer), epigenetic ageing was estimated recently to occur at a faster rate in the active season compared to the hibernation season [21].
Hibernation occurs in many species of bats found in temperate regions [3,4,22] and is of particular interest because it is associated with the evolution of extreme longevity [23]. Here, we used big brown bats, Eptesicus fuscus, to investigate the effect of hibernation on patterns of DNAm by microarray profiling over 37 000 conserved CpG sites [24]. Many of these sites exhibit reliable change in DNAm with age and, as a consequence, DNAm can be used to accurately predict chronological age in many bat species [25]. In addition, some sites show differences between short and long-lived species in the rate of change of DNAm [25], suggesting that DNAm profiles can provide information regarding the rate of ageing, as has been demonstrated for humans [26–28]. To assess the influence of hibernation on DNAm profiles, we used tissue samples taken in the winter and summer from bats that undergo natural annual hibernation. We used these data to determine if epigenetic age, as predicted by a multi-species bat epigenetic clock, differs between summer and winter samples, and to identify sites in the genome that exhibit change in DNAm associated with hibernation independent of age. Given the results of prior gene expression studies, we hypothesize that animals experience reduced epigenetic ageing during hibernation and that CpG sites that exhibit change in DNAm during hibernation are near genes associated with metabolism and immunity. In addition, we test if CpG sites differentially methylated and associated with hibernation are independent of sites associated with differences in bat longevity [25].
2. Methods
The E. fuscus sampled in this study were housed in a captive research colony at McMaster University in Hamilton, Ontario, Canada (43.26° N, −79.92° W). Bats in the colony live in an indoor free-flight area (2.5 × 1.5 × 2.3 m) with year-round access to a larger (2.5 × 3.8 × 2.7 m) outdoor flight enclosure [29]. Colony temperature and lighting varied with ambient conditions; however, in the winter, a heater kept the indoor colony temperature above 6°C. Bats have ad libitum access to water and food (yellow mealworms, Tenebrio molitor, Reptile Feeders, Norwood, ON). Minimum ages were available for six animals caught as adults 6–9 years before sampling and exact ages for 14 animals born in captivity. When first sampled, these 20 animals ranged from 0.7 to 10.1 years of age. Twelve bats were sampled in February 2020, 20 bats in July 2020 and seven additional bats in February 2021. We excluded one sample from five of the wild-caught animals because of illness at sampling, which left 15 matched pairs from the same animal (12 winter–summer, 3 summer–winter). Big brown bats in Ontario typically initiate hibernation in late October and leave hibernation in early April although animals periodically arouse every 30–45 days throughout the winter to drink and eliminate metabolic waste. Thus, winter samples were taken from captive animals during the hibernation period and summer samples were from the active period.
We used a circular, sterile Sklar Tru-Punch biopsy tool to excise one or two 3-mm diameter tissue biopsies from the wing membrane of each bat. Genomic DNA was extracted from wing tissue using a Zymo Quick-DNA miniprep plus kit (ZymoResearch, Orange, CA, USA) using the standard tissue protocol. Samples with at least 250 ng of DNA but with a concentration below 10 ng µl−1 were concentrated using a 30 kDa centrifugal filter to reach at least 12.5 ng µl−1 in 20 µl (MilliporeSigma, Burlington, MA, USA).
DNA samples were processed at the UCLA Neuroscience Genomics core facility to measure DNAm. After bisulfite conversion, samples were hybridized to a custom Illumina microarray (HorvathMammalMethylChip40) containing 37 492 probes of conserved 50 base pair sequences with terminal CpG sites. The microarray was designed to assay DNAm from any mammal using probes largely conserved across 62 mammal species [24]. Alignment of the microarray probe sequences to the E. fuscus genome (v. 1.0) identified genomic positions for 32 217 CpG sites [25]. These sites were categorized depending on the nearest transcription start site (TSS) for annotated genes as being in either intergenic, 3′ UTR (untranslated region), 5′ UTR, promoter (minus 10 kb to plus 1000 bp from the nearest TSS), exon or intron regions. The proportion of DNA molecules methylated at each CpG site (i.e. Beta values) were obtained after normalization using the SeSaME procedure [30] to correct for bias or variation among plates.
We tested if hibernation influences epigenetic ageing by using a multi-species epigenetic clock created for 712 known-aged bats from 26 species [25]. As noted above, we had minimum ages for 6 of 20 bats (30%). We independently estimated the ages of these animals using a species-specific epigenetic clock created using ‘glmnet’ in R [31], which fitted an elastic net regression model using an alpha of 0.5 between DNAm beta values and chronological age for 60 previously profiled, known-aged E. fuscus [25]. The estimated ages were, on average, within 1 year of the minimum ages. We then calculated the residuals between the multi-species clock predicted age and the chronological age of each individual, and compared those residuals between winter (hibernating) and summer (non-hibernating) samples using a linear mixed model, with season and birthplace (captive or wild) as fixed effects and bat identity as a random effect fitted by restricted maximum likelihood. Summary data are reported as mean ± standard error.
To determine if hibernation influences DNAm independent of age at each CpG site, we fitted linear models for DNAm on age using the lm function in R [32] for all E. fuscus samples. We then used the resulting residuals in a linear mixed model to compare winter and summer age-adjusted DNAm measured from the same individual with individual included as a random effect. To correct for multiple testing, we used a Benjamini–Yekutieli (BY) false discovery rate (FDR) to obtain adjusted p-values [33]. Significant CpG sites (BY p < 0.05) are referred to as differentially methylated positions (DMPs). Because this analysis compared age-adjusted DNAm in winter to summer, sites with higher DNAm in the winter are referred to as ‘winter-up’ and sites with lower DNAm in the winter are referred to as ‘winter-down’ (see figure 2 for examples).
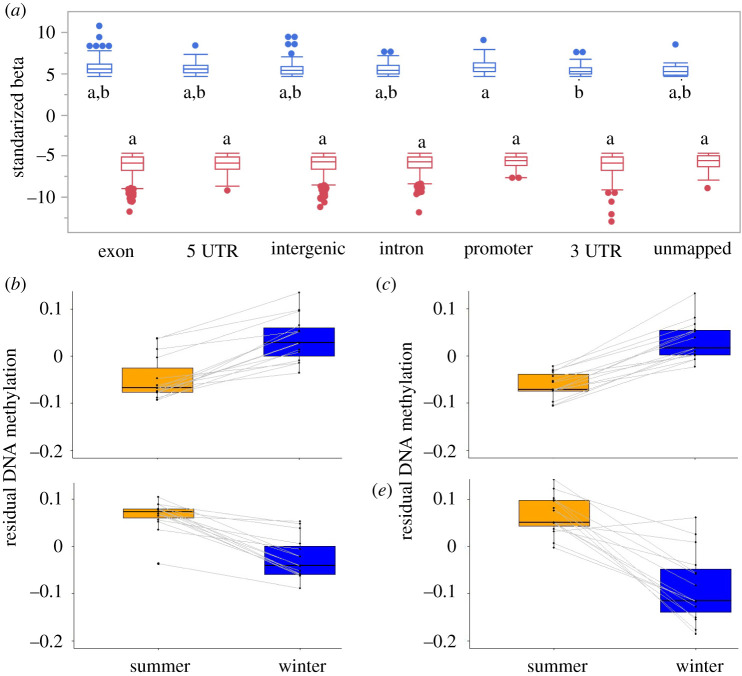
Figure 2.
(a) Magnitude of the effect of hibernation on significant DMPs among genomic regions, as indicated by box plots showing median, quartiles, whiskers (1.5 times the interquartile range) and outliers of the standardized beta coefficient. Winter-up DMPs in blue, winter-down DMPs in red. Letters indicate results of a Tukey's post hoc test with alpha = 0.05. Example winter-up DMPs include (b) cg10775708 in an intergenic region near MAZ (MYC-associated zinc finger protein) and (c) cg05808663 in the promoter region of NFIX (Nuclear Factor I X). Example winter-down DMPs include (d) cg03445006 in an intergenic region near BNC2 (Basonuclin 2) and (e) cg01905984 in the promoter region of PHACTR2 (Phosphatase and Actin Regulator 2). In (b–e), lines connect samples from the same individual. (Online version in colour.)
We used contingency tests to determine if winter-up or winter-down DMPs were non-randomly distributed in the genome relative to genomic regions and conducted an analysis of variance on the standardized regression coefficients to determine if the magnitude of the effect differed among genomic regions. We compared distance to TSSs between significant and nonsignificant sites using a Student's t-test. We also conducted enrichment analyses on DMPs using experimentally derived functional element overlap analysis of ReGions from EWAS, eFORGE v. 2.0 [34]. This programme tests for enrichment of functional regulatory elements by identifying CpG probe sequences associated with DNAse hypersensitive sites, each of five different histone marks or 15 inferred chromatin states identified in human cell lines derived from multiple tissue sources by the Epigenomics Roadmap Consortium (https://egg2.wustl.edu/roadmap/web_portal/). By comparing DMPs to chromatin state or histone associations in human cell lines, eFORGE allows for tissue-specific enrichment tests of DNAm. Permutation tests are then run against the species' genomic background to determine which functional elements occur non-randomly [34]. We conducted separate analyses for winter-up and winter-down DMPs only using sites with an adjusted p-value below 0.05 or the most significant 1000 sites in each direction if there were more than 1000 significant sites. As background for the enrichment analysis we selected E. fuscus with a 1 kb proximity window. These options constrain the analysis to the 32 217 sites on the 37 K array that have been mapped to the E. fuscus genome and use only one site within 1000 bp on each of 1000 permutations. As bat wing tissue is comprised of skin, muscle and blood [35], we only report results for the cell lines derived from similar tissues. For the histone mark analysis there were 7 blood cell lines, 3 fetal muscle cell lines and 4 foreskin cell lines while the chromatin state analysis used 27 blood cell lines, 12 muscle cell lines and 10 skin cell lines. We used a BY FDR of 1% to identify enriched regulatory elements.
We then conducted several association tests to determine the function of genes closest to hibernation DMPs. As multiple probes on the array often are located nearest the same gene, the DNAm effect direction on each gene was determined based on the number of winter-down or winter-up DMPs. Genes with a greater number of significant winter-down sites were considered to have reduced DNAm in winter and genes with a greater number of winter-up sites were considered to have elevated DNAm in winter. Genes with an equal number of winter-up and winter-down DMPs were not considered to have a significant directional effect. We considered all 4808 known genes nearest to mapped CpG sites in the E. fuscus genome annotation as background and then used PANTHER v. 16.0 [36] to test for enrichment of winter-up and winter-down genes with respect to protein class, cellular component or biological process using a Fisher's exact test (FET, FDR < 0.05). In a prior study using the same array platform, a total of 1491 CpG sites (corresponding to 700 E. fuscus genes) exhibited significant differences in the rate of DNAm change between three long-lived and two short-lived bat species [25]. We used FETs to compare winter-up and winter-down genes to this list of 700 longevity genes and to 4723 innate immunity genes (downloaded from https://www.innatedb.com, 14 August 2020) to determine if hibernation genes are also involved in innate immunity.
3. Results
(a) . Hibernation slows epigenetic ageing
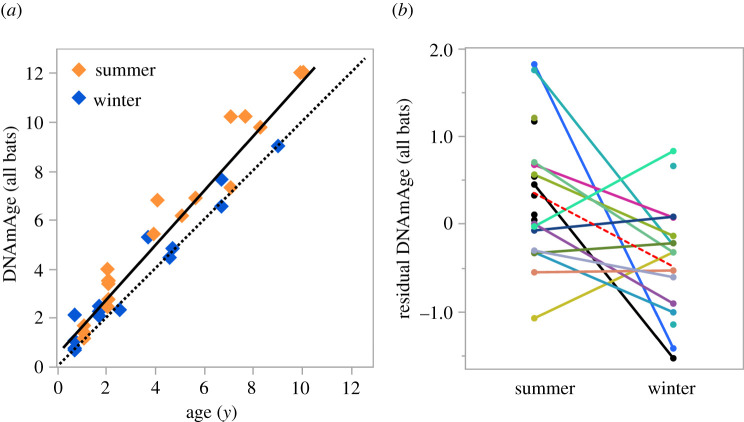
A multi-species epigenetic clock [25] predicted ages that correlate highly with the chronological ages of big brown bats (r2 = 0.941, median absolute error (MAE) = 0.531), although the predicted age is equal to or greater than the chronological age in most cases (figure 1a). Moreover, the residuals from a regression of predicted epigenetic age on chronological age differ between summer and winter samples (F = 7.94, d.f. = 1, 23, p = 0.0097), but not birthplace (F = 0.13, d.f. = 1, 30, p = 0.72), and reveal that the epigenetic age of hibernating bats is 0.77 ± 0.25 years less than the epigenetic age of non-hibernating bats (figure 1b).
Figure 1.
Multi-species epigenetic clock applied to E. fuscus sampled in summer and winter. (a) Least-squares fit (solid line) between chronological age and biological age as predicted by a 26 bat species epigenetic clock from Wilkinson et al. [25] with the predicted chronological age indicated by the dashed y = x line. (b) Residuals from the regression in (a) plotted for each non-hibernating (summer) and hibernating (winter) bat. Solid lines connect individuals with wild-born animals labelled black. The dashed line indicates the change in mean values across seasons. (Online version in colour.)
(b) . Hibernation primarily increases DNAm across the genome
A total of 3002 CpG sites mapped (and 78 unmapped) in the E. fuscus genome showed significant differences in age-adjusted DNAm across seasons with significantly more winter-up (77.5%) than winter-down DMPs (X2 = 415.5, p = 2.34 × 10−92). Hibernation DMPs were widely distributed on 195 of 361 genomic scaffolds but not distributed at random with respect to genomic region (X2 = 336.7, p = 3.28 × 10−75). Significant DMPs were further from TSSs than nonsignificant sites in absolute base pair distance (112 021 ± 2879 versus 82 334 ± 7966 bp; t = 9.94, p < 0.0001), but winter-down DMPs were closer than winter-up DMPs (85 607 ± 5445 versus 119 712 ± 3347 bp; t = 5.33, p < 0.0001) to TSSs. Corresponding to those genomic locations, more DMPs than expected were located in introns (908 observed/711.8 expected = 1.28; X2 = 54.1, p = 1.92 × 10−13) and intergenic regions (956/698.5 = 1.37; X2 = 95.0, p = 1.94 × 10−22, but fewer DMPs than expected were located in exons (803/1024.6 = 0.78; X2 = 47.9, p = 4.44 × 10−12), promoter regions (119/202.7 = 0.59; X2 = 34.5, p = 4.18 × 10−9) and 5′UTRs (108/238.3 = 0.45; X2 = 71.2, p = 3.21 × 10−17). DMPs in 3′UTR regions were not enriched (108/126.2 = 0.86; X2 = 2.64, p = 0.10). By contrast, the magnitude of the effect due to the difference in residual DNAm between winter and summer at DMPs showed only minor variation among genomic regions at sites with either decreased (F = 2.14, d.f. = 6,687, p = 0.047) or increased (F = 3.33, d.f. = 6 2379, p = 0.003) methylation during winter with the only significant difference revealed by post hoc tests occurring between winter-up sites at promoters and 3′UTR regions (figure 2a).
(c) . DNAm changes associated with hibernation influence metabolic processes, immunity and longevity
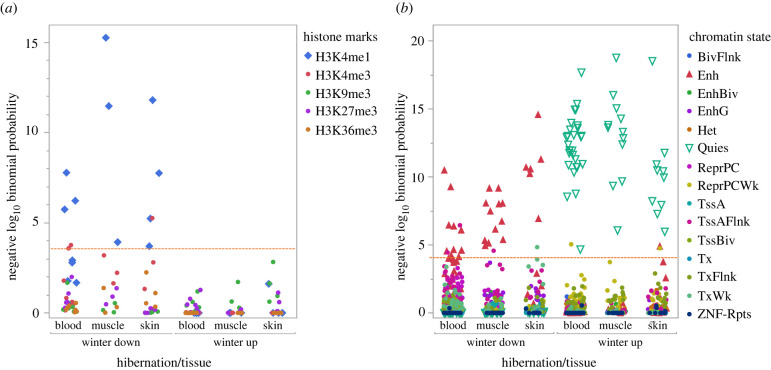
An eFORGE analysis revealed that one histone mark, H3K4me1, which is often associated with active or primed enhancers, was enriched in multiple cell lines from all three tissue sources in winter-down DMPs, but no histone mark was significantly associated with winter-up DMPs (figure 3a). The eFORGE analysis of inferred chromatin states using winter-down DMPs also indicates significant enrichment for enhancers in cell lines derived from blood, skin and muscle tissues while winter-up DMPs were highly enriched for quiescent states in cell lines from all three tissue sources (figure 3b).
Figure 3.
Evidence of tissue-specific epigenomic enrichment obtained from eFORGE for (a) 5 histone marks across 7 blood cell lines, 3 muscle cell lines and 4 skin cell lines and (b) 15 chromatin states across 14 blood cell lines, 6 muscle cell lines and 8 skin cell lines. Chromatin state abbreviations are BivFlnk, flanking bivalent TSS/Enh; Enh, enhancers; EnhBiv, bivalent enhancer; EnhG, genic enhancers; Het, heterochromatin; Quies, quiescent/low; ReprPC, repressed PolyComb; ReprPCWk, weak repressed PolyComb; TssA, active TSS; TssAFlnk, flanking active TSS; TssBiv, bivalent/poised TSS; Tx, strong transcription; TxFlnk, transcription at gene 5′ and 3′; TxWk, weak transcription; ZNF/Rpts, zinc finger genes and repeats. Red dashed line indicates significance (BY adjusted p < 0.01). (Online version in colour.)
Winter-up DMPs were nearest 739 genes and winter-down DMPs were nearest 219 genes. Gene ontology enrichment tests for cellular components revealed that genes associated with chromatin were significantly enriched for winter-up and winter-down genes. Winter-down genes were also significantly enriched for nuclear, chromosomal and non-membrane-bounded organelle cell components. Winter-down and winter-up genes were enriched for gene specific transcription regulators and transcription factors (figure 4a). Indeed, over 40 biological processes were significant after adjusting for multiple testing. The top 15 significant biological processes were associated with winter-down genes and 13 of those were associated with regulation of metabolism or transcription (figure 4b).
Figure 4.
Gene ontology enrichment analysis for (a) protein class and (b) biological process. Significance from a Fisher's exact rest is indicated by colour in each cell. The threshold 1% FDR is –log10 p = 3.5. (c) Number and overlap of genes nearest significant DMPs associated with hibernation, innate immunity and longevity. (Online version in colour.)
Significant overlap was also found between innate immunity genes and genes nearest hibernation DMPs (figure 4c). Of the 1071 innate immunity genes with sites on the array, 248 (23.2%) exhibited evidence of differential methylation (X2 = 9.02, p = 0.0027), a majority of which (188; 75.8%) were associated with elevated DNAm during hibernation. Comparison of 700 previously identified bat longevity genes to the 958 hibernation genes reveals 306 genes in common (figure 4c)—dramatically more than expected by chance (X2 = 290.6, p = 3.68 × 10−65), with 52 (17%) longevity genes associated with winter-down DMPs. Moreover, 41 genes nearest winter-down DMPs and associated with bat longevity exhibit differential gene expression during hibernation or torpor in other mammals and six have been previously identified as candidate torpor genes (electronic supplementary material, table S1).
4. Discussion
Application of a multi-species bat epigenetic clock [25] provides strong evidence that hibernation is associated with slower epigenetic ageing. The multi-species clock explains 94% of the variation in the chronological ages of both hibernating and non-hibernating big brown bats; however, the clock estimates are equal to or greater than the chronological age, suggesting big brown bats age slightly faster than a ‘typical’ bat, especially during the active period (figure 1). The multi-species bat clock was created using DNAm data from 26 species, some of which have longer lifespans for their body sizes than big brown bats (cf. figure 2, [25]). Moreover, comparison of deviations from a regression of predicted on true ages reveals that the epigenetic age of hibernating bats is nearly a year less than non-hibernating bats after adjusting for age. This finding provides direct evidence that hibernation is associated with reduced epigenetic ageing in bats (Order Chiroptera), and is consistent with a recent study in marmots (Order Rodentia) that reached the same conclusion [21]. Because the multi-species clock only uses DNAm from 162 CpG sites, this result does not reveal the extent to which hibernation influences DNAm in the genome. After adjusting for age, comparison of DNAm at over 32 000 sites profiled in samples taken from the same animal during the winter hibernation period versus the summer active season reveals that hibernation alters DNAm at over 3000 DMPs widely distributed in the genome of E. fuscus. Over three times as many DMPs exhibited increased DNAm as opposed to decreased DNAm during hibernation. Given that elevated DNAm is typically associated with suppressed gene transcription, this result is consistent with a reduction in the activity of many genes during hibernation.
The amount of DNAm can vary by tissue; however, the eFORGE analysis revealed evidence of consistent enrichment of histone marks and inferred chromatin states in multiple human cell lines derived from blood, skin and muscle tissues (figure 3). Winter-up DMPs were not associated with any histone marks but were highly associated with quiescent chromatin states in all cell lines. Given that quiescence is a stress-resistant state in which cells suspend most processes, this finding is consistent with reduced metabolism and immune function during hibernation [6,37]. By contrast, winter-down DMPs were enriched for H3K4me1, a histone mark associated with enhancers [38] and enriched for enhancer chromatin states. In other words, winter-down DMPs were associated with active gene transcription. We suspect these results provide a conservative estimate of the number of regulatory factors involved both because the array only samples a fraction of possible enhancer regions and because enhancer motifs can differ among mammals [39]. Nevertheless, the strength and consistency of the evidence for enhancer enrichment across multiple cell lines indicates there must be substantial sequence conservation at these regions.
Enrichment analysis of winter-up genes failed to detect significant enrichment for any biological process, indicating that elevated DNAm during hibernation is associated with a diverse ensemble of genes in proportion to their relative abundance on the array. By contrast, enrichment analysis of winter-down genes found evidence for enrichment of DNA-binding transcription factors, especially those involved in regulating metabolic processes (figure 4). This result is consistent with the eFORGE analysis and with previous findings on squirrels and marsupials that found increased expression of genes involved in cold-tolerant mechanisms of metabolism and regulation of pathways involved in protein turnover [5,40] or cryoprotection [5,41] during hibernation.
In addition to being associated with regulating metabolic processes, genes nearest to hibernation DMPs were also more likely to be involved in innate immunity and bat longevity as indicated by different age-dependent rates of change in DNAm between short-lived and long-lived bats [25]. Most (79%) of the 52 genes nearest winter-down DMPs associated with bat longevity exhibit differential gene expression during torpor in other mammals (electronic supplementary material, table S1). These results are consistent with longevity in bats being associated with periodic episodes of reduced energy consumption due to lowered metabolism and suppressed immune function. This study illustrates how DNAm profiling using nonlethal tissue samples can provide real-time insight into the epigenetic control of physiological processes, such as hibernation.
Acknowledgements
We thank S. Horvath and J. Zhang for data normalization and analysis, K. Delaney and D. Graham for veterinary support and animal care, and K. Bennett, P. Johnson, S. Mount, K. Paczolt and three anonymous reviewers for comments.
Ethics
All experimental procedures met the Guide to the Care and Use of Experimental Animals published by the Canadian Council on Animal Care and were approved by the Animal Research Ethics Board of McMaster University.
Data accessibility
All data used in this study are freely available. Normalized methylation values for each sample, along with sample metadata, are available from NCBI as series GSE164127 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164127). The design of the Illumina microarray (HorvathMammalMethylChip40) is available from NCBI as platform GPL28271 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL28271). R scripts, clock coefficients, age predictions, probe annotations for the E. fuscus genome, eFORGE output and statistics for identifying DMPs are available at doi:10.6084/m9.figshare.c.5927288.v1 [42].
Electronic supplementary material is available online [43].
Authors' contributions
I.R.S.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, software, writing—original draft, writing—review and editing; D.M.A.: conceptualization, data curation, formal analysis, investigation, methodology, software, writing—original draft, writing—review and editing; L.J.S.G.: data curation, methodology, resources, writing—review and editing; P.A.F.: data curation, funding acquisition, methodology, resources, supervision, writing—review and editing; G.S.W.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Funding was provided by the American Society of Mammalogists, Sigma Xi, the University of Maryland Graduate Student Government, the College of Computer, Math and Natural Sciences, Discovery grant no. RGPIN-2020-06906 to P.A.F. from the Natural Sciences and Engineering Research Council (NSERC) of Canada and a NSERC Post-Graduate Scholarship to L.J.S.G.
References
- 1.Jastroch M, Giroud S, Barrett P, Geiser F, Heldmaier G, Herwig A. 2016. Seasonal control of mammalian energy balance: recent advances in the understanding of daily torpor and hibernation. J. Neuroendocrinol. 28. ( 10.1111/jne.12437) [DOI] [PubMed] [Google Scholar]
- 2.Ruf T, Geiser F. 2015. Daily torpor and hibernation in birds and mammals. Biol. Rev. Camb. Philos. Soc. 90, 891-926. ( 10.1111/brv.12137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czenze ZJ, Park AD, Willis CKR. 2013. Staying cold through dinner: cold-climate bats rewarm with conspecifics but not sunset during hibernation. J. Comp. Physiol. B 183, 859-866. ( 10.1007/s00360-013-0753-4) [DOI] [PubMed] [Google Scholar]
- 4.Turbill C, Bieber C, Ruf T. 2011. Hibernation is associated with increased survival and the evolution of slow life histories among mammals. Proc. R. Soc. B 278, 3355-3363. ( 10.1098/rspb.2011.0190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nespolo RF, Gaitan-Espitia JD, Quintero-Galvis JF, Fernandez FV, Silva AX, Molina C, Storey KB, Bozinovic F. 2018. A functional transcriptomic analysis in the relict marsupial Dromiciops gliroides reveals adaptive regulation of protective functions during hibernation. Mol. Ecol. 27, 4489-4500. ( 10.1111/mec.14876) [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto M, Gao B, Kikuchi-Utsumi K, Ohinata H, Osborne PG. 2002. Arousal from hibernation and BAT thermogenesis against cold: central mechanism and molecular basis. J. Therm. Biol. 27, 503-515. ( 10.1016/S0306-4565(02)00024-4) [DOI] [Google Scholar]
- 7.Hindle AG, Martin SL. 2013. Intrinsic circannual regulation of brown adipose tissue form and function in tune with hibernation. Am. J. Physiol. Endocrinol. Metab. 306, E284-E299. ( 10.1152/ajpendo.00431.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y-H, Zhang Y, Cui J, Liu Y, McAllan BM, Liao C-C, Zhang S. 2013. Adaptation of phenylalanine and tyrosine catabolic pathway to hibernation in bats. PLoS ONE 8, 4. ( 10.1371/journal.pone.0062039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CW, Biggar KK, Luu BE, Szereszewski KE, Storey KB. 2016. Analysis of microRNA expression during the torpor-arousal cycle of a mammalian hibernator, the 13-lined ground squirrel. Physiol. Genom. 48, 388-396. ( 10.1152/physiolgenomics.00005.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorov VB, et al. 2014. Comparative functional genomics of adaptation to muscular disuse in hibernating mammals. Mol. Ecol. 23, 5524-5537. ( 10.1111/mec.12963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessier SN, Storey KB. 2014. To be or not to be: the regulation of mRNA fate as a survival strategy during mammalian hibernation. Cell Stress Chaperones 19, 763-776. ( 10.1007/s12192-014-0512-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Attar R, Storey KB. 2020. Suspended in time: molecular responses to hibernation also promote longevity. Exp. Gerontol. 134, 110889. ( 10.1016/j.exger.2020.110889) [DOI] [PubMed] [Google Scholar]
- 13.Grabek KR, Martin SL, Hindle AG. 2015. Proteomics approaches shed new light on hibernation physiology. J. Comp. Physiol. B 185, 607-627. ( 10.1007/s00360-015-0905-9) [DOI] [PubMed] [Google Scholar]
- 14.Alvarado S, Mak T, Liu S, Storey KB, Szyf M. 2015. Dynamic changes in global and gene-specific DNA methylation during hibernation in adult thirteen-lined ground squirrels, Ictidomys tridecemlineatus. J. Exp. Biol. 218, 1787-1795. ( 10.1242/jeb.116046) [DOI] [PubMed] [Google Scholar]
- 15.Biggar Y, Storey KB. 2014. Global DNA modifications suppress transcription in brown adipose tissue during hibernation. Cryobiology 69, 333-338. ( 10.1016/j.cryobiol.2014.08.008) [DOI] [PubMed] [Google Scholar]
- 16.Moore LD, Le T, Fan G. 2013. DNA methylation and its basic function. Neuropsychopharmacology 38, 23-38. ( 10.1038/npp.2012.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade PA. 2001. Methyl CpG-binding proteins and transcriptional repression. BioEssays 23, 1131-1137. ( 10.1002/bies.10008) [DOI] [PubMed] [Google Scholar]
- 18.Rose NR, Klose RJ. 2014. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 1839, 1362-1372. ( 10.1016/j.bbagrm.2014.02.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razin A, Riggs AD. 1980. DNA methylation and gene function. Science 210, 604-610. ( 10.1126/science.6254144) [DOI] [PubMed] [Google Scholar]
- 20.Fujii G, Nakamura Y, Tsukamoto D, Ito M, Shiba T, Takamatsu N. 2006. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem. J. 395, 203-209. ( 10.1042/BJ20051802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinho GM, et al. 2022. Hibernation slows epigenetic ageing in yellow-bellied marmots. Nat. Ecol. Evol. 6, 418-426. ( 10.1038/s41559-022-01679-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blažek J, et al. 2019. Numerous cold arousals and rare arousal cascades as a hibernation strategy in European Myotis bats. J. Therm. Biol. 82, 150-156. ( 10.1016/j.jtherbio.2019.04.002) [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson GS, Adams DM. 2019. Recurrent evolution of extreme longevity in bats. Biol. Lett. 15, 20180860. ( 10.1098/rsbl.2018.0860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arneson A, et al. 2022. A mammalian methylation array for profiling methylation levels at conserved sequences. Nat. Commun. 13, 783. ( 10.1038/s41467-022-28355-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson GS, et al. 2021. DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun. 12, 1615. ( 10.1038/s41467-021-21900-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S, Raj K. 2018. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371-384. ( 10.1038/s41576-018-0004-3) [DOI] [PubMed] [Google Scholar]
- 27.Marioni RE, et al. 2015. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16, 25. ( 10.1186/s13059-015-0584-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. 2016. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenetics 8, 64. ( 10.1186/s13148-016-0228-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skrinyer AJ, Faure PA, Dannemiller S, Ball HC, Delaney KH, Orman R, Stewart M, Cooper LN. 2017. Care and husbandry of the world's only flying mammals. Lab. Anim. Sci. Prof. June, 24-27. [Google Scholar]
- 30.Zhou W, Triche TJ Jr, Laird PW, Shen H. 2018. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, e123. ( 10.1093/nar/gky691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1-22. ( 10.18637/jss.v033.i01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Team R.C. 2020. R: a language and environment for statistical computing. (4.0.2 ed). Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 33.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165-1188. ( 10.1214/aos/1013699998) [DOI] [Google Scholar]
- 34.Breeze CE, et al. 2019. eFORGE v2.0: updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 35, 4767-4769. ( 10.1093/bioinformatics/btz456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheney JA, Allen JJ, Swartz SM. 2017. Diversity in the organization of elastin bundles and intramembranous muscles in bat wings. J. Anat. 230, 510-523. ( 10.1111/joa.12580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi H, Ebert D, Muruganujan A, Mills C, Albou L-P, Mushayamaha T, Thomas PD. 2020. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394-D403. ( 10.1093/nar/gkaa1106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouma HR, Carey HV, Kroese FG. 2010. Hibernation: the immune system at rest? J. Leukocyte Biol. 88, 619-624. ( 10.1189/jlb.0310174) [DOI] [PubMed] [Google Scholar]
- 38.Rada-Iglesias A. 2018. Is H3K4me1 at enhancers correlative or causative? Nat. Genet. 50, 4-5. ( 10.1038/s41588-017-0018-3) [DOI] [PubMed] [Google Scholar]
- 39.Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160, 554-566. ( 10.1016/j.cell.2015.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermillion KL, Anderson KJ, Hampton M, Andrews MT. 2015. Gene expression changes controlling distinct adaptations in the heart and skeletal muscle of a hibernating mammal. Physiol. Genom. 47, 58-74. ( 10.1152/physiolgenomics.00108.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szereszewski KE, Storey KB. 2019. Identification of a prosurvival neuroprotective mitochondrial peptide in a mammalian hibernator. Cell Biochem. Funct. 37, 494-503. ( 10.1002/cbf.3422) [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson G, Adams D, Sullivan I. 2022. Supporting information for “Big brown bats experience slower epigenetic ageing during hibernation”. Figshare. ( 10.6084/m9.figshare.c.5927288.v1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan IR, Adams DM, Greville LJS, Faure PA, Wilkinson GS. 2022. Big brown bats experience slower epigenetic ageing during hibernation. Figshare. ( 10.6084/m9.figshare.c.6123995) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study are freely available. Normalized methylation values for each sample, along with sample metadata, are available from NCBI as series GSE164127 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164127). The design of the Illumina microarray (HorvathMammalMethylChip40) is available from NCBI as platform GPL28271 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL28271). R scripts, clock coefficients, age predictions, probe annotations for the E. fuscus genome, eFORGE output and statistics for identifying DMPs are available at doi:10.6084/m9.figshare.c.5927288.v1 [42].
Electronic supplementary material is available online [43].