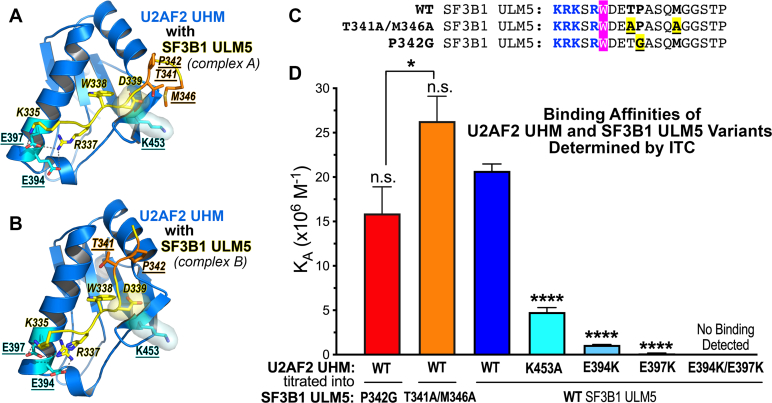
Figure 3.
Contribution of interface residues to the U2AF2 UHM–SF3B1 ULM5 binding affinity.A and B, interactions mediated in the two copies of the U2AF2 UHM (marine)–SF3B1 ULM5 (yellow, italicized labels) complex tested by structure-guided mutagenesis (underlined labels; U2AF2: cyan; SF3B1: orange). C, sequences of the SF3B1 ULM5 peptides (residues 333–351) showing the structure-guided mutants (yellow) tested by isothermal titration calorimetry. The central tryptophan is magenta and the basic residues are blue. D, bar graph of the average apparent binding affinities (KA) and standard deviations of three isothermal titration calorimetry experiments. The thermodynamic values and representative isotherms are given Table S1 and Fig. S1. The significance of the differences between the mutant and WT affinities are given, except where indicated by lines between the P342G and T341A/M346A pair; n.s., not significant, p > 0.05; ∗p < 0.05; ∗∗∗∗p < 0.0005.