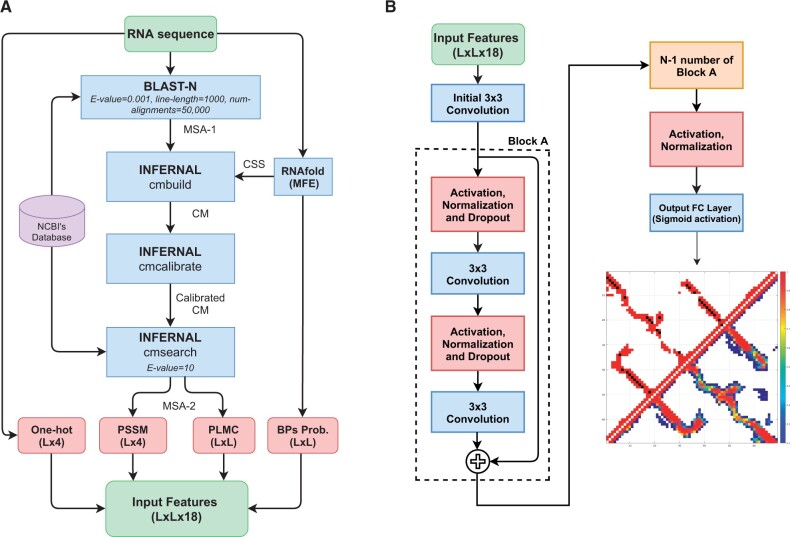
Fig. 1.
(A) Inputted 1D and 2D features used by the SPOT-RNA-2D; where MSA is multiple sequence alignment, CSS is predicted consensus secondary structure from RNAfold (MFE), CM is covariance model, One-hot is the one-hot encoding of the input sequence, PSSM is the position-specific scoring matrix, PLMC is pseudo-likelihood maximization coupling, BPs is predicted base-pairs probability from RNAfold (MFE), MFE is minimum free energy and L is the length of the RNA sequence. (B) The generalized deep neural network architecture of SPOT-RNA-2D