Fig. 2. A cartoon describing conserved secondary and tertiary structures among PI3Kγ, PKA, IPMK and IP3KA.
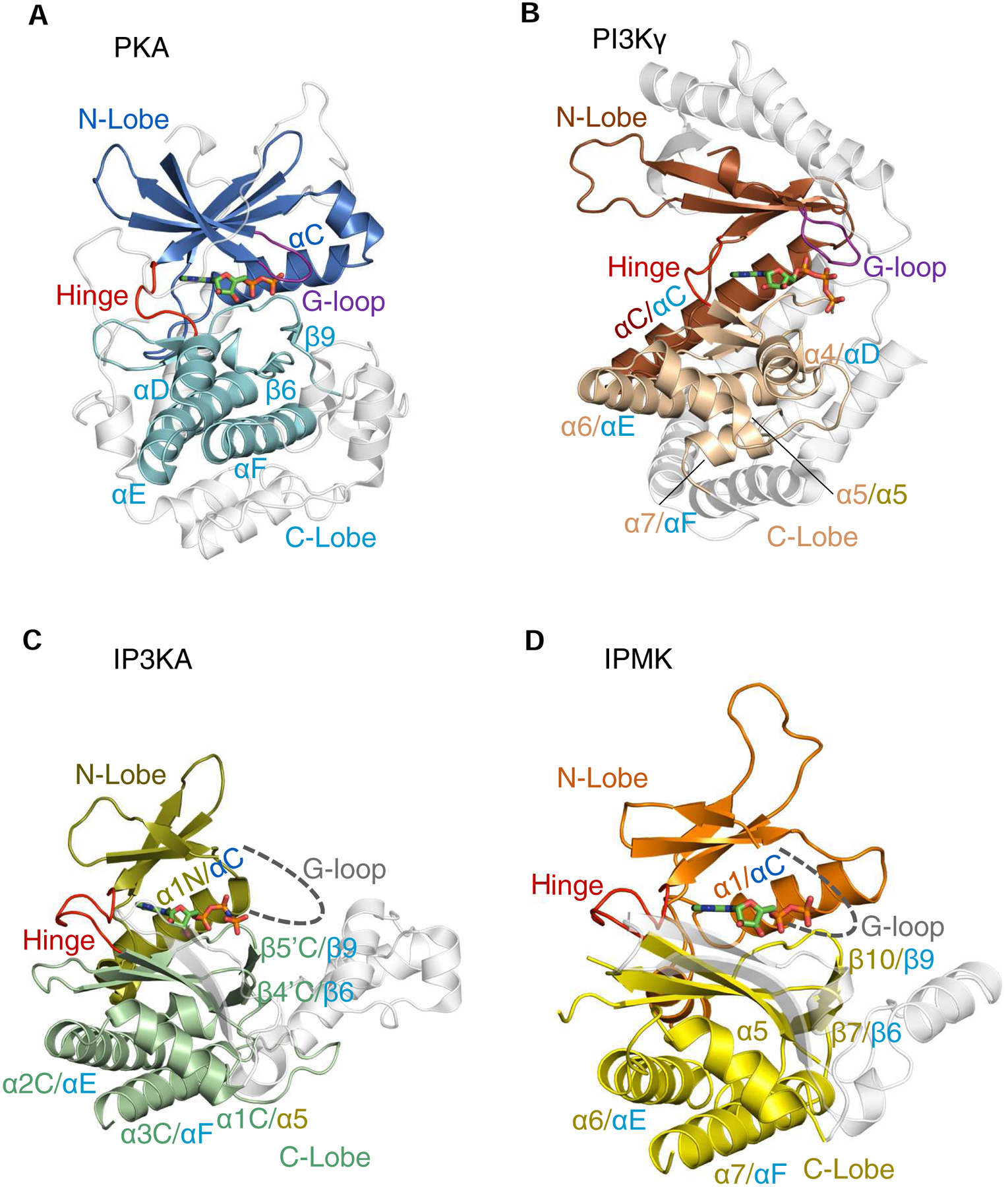
Ribbon plots of the following enzymes are shown (PDB access codes in parentheses): A, Murine protein kinase A (catalytic subunit)/ADP complex, E.C. 2.7.11.11 (1L3R); B, Sus scrofa PI3Kγ (catalytic subunit)/ATP complex (E.C. 2.7.11.1) (1E8X). C, Human IP3KA/AMP-PNP complex (1W2C) D, Human IPMK/ADP/IP3 complex (5W2H). The α-helices and β-strands that are colored are conserved in at least two of the structures shown; in cases where individual structural elements have two labels separated by a forward slash, the first is the label given in the original PDB entry, and the second label corresponds to the corresponding element in the homologous protein (color coding of labels matches that of the originating lobe). Non-conserved structural elements are depicted in light gray. Broken lines are used to depict the expected G-loops in IP3K and IPMK (their actual structures are not known). Bound nucleotides are shown as stick models. Note that the structural element in PI3Kγ that we describe as a ‘G-loop’ (panel B) was originally named ‘P-loop’ (Walker et al., 1999). For an explanation, see the text.
