Fig. 4. Cooperation between the lobes: R-Spines, gatekeepers, catalytic triads and other motifs.
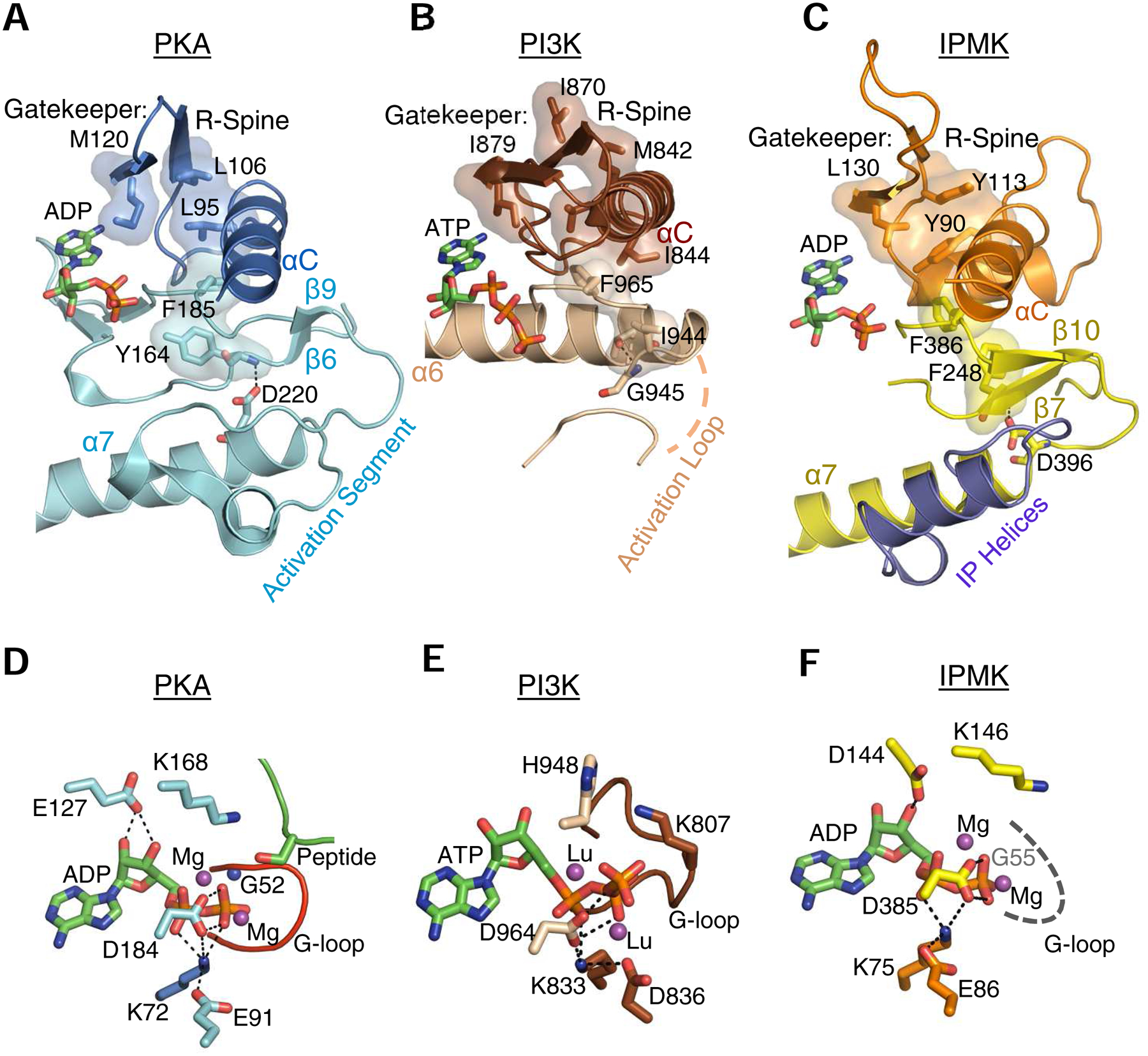
A, B and C highlight selected, conserved elements of murine PKA (PDB, 1L3R), human IPMK (PDB, 5W2H) and Sus scrofa PI3Kγ (PDB, 1E8X), respectively, shown as a composite of ribbon plots, with selected key residues as space-filling representations and stick-model depictions. For PKA and PI3Kγ, the identities of residues comprising the gatekeeper, R-spine and the anchoring α-helix are provided in reviews of this topic (Taylor and Kornev, 2011; Vadas et al., 2011); the corresponding features in IPMK were identified from its alignment with PKA (using Pymol) in which bound nucleotides were superimposed. Panels D, E, F are corresponding stick-model close-ups of the catalytic pockets. The dashed gray line in panel F denotes that the actual structure of the G-loop is disordered in the crystal structure. The residues that we denote as comprising a ‘catalytic triad’ (see text) are as follows: PKA, Lys72, Glu91, Asp184; PI3Kγ, Lys833, Asp836, Asp964; IPMK, Lys75, Glu86, Asp385 (also see Table 1). Thin, broken black lines depict interactions between residues in the catalytic triad, and also those involving the nucleotide’s ribose ring. Magenta balls depict the metal binding sites occupied by magnesium (or, in the case of PI3Kγ, lutetium (Walker et al., 1999)). All other color schemes match those in Fig. 2. Note that the structural element in PI3Kγ that we describe as a ‘G-loop’ (panel E) was originally named ‘P-loop’ (Walker et al., 1999). For an explanation, see the text.
