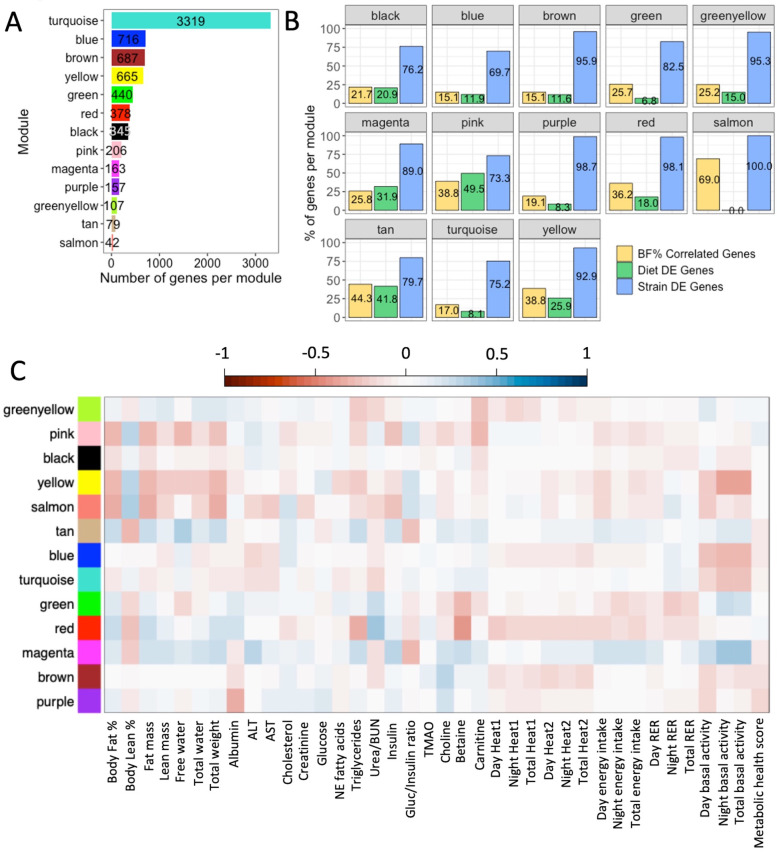
Fig. 5.
WGCNA identified co-regulated gene modules correlated with phenotypic traits. Using the cleaned and filtered hepatic gene expression data from mice fed the HP diet and mice fed the HS diet, A weighted gene co-expression network analysis (WGCNA) identified 13 modules with arbitrarily assigned colors. The 11,542 expressed genes were used to form the modules, which varied widely in terms of the number genes within each module. Gene module assignments are shown in Supplementary Table 9, Additional file 2. B Modules demonstrated a wide compositional range in terms of genes with expression levels significantly correlated with body fat % (15.1–69.0%) and differential expression by diet (0–49.5%) but consistently contained a high proportion of genes differentially expressed by CC strain (69.7–100%). C The heatmap of Spearman’s correlations between module eigengenes and phenotypic traits measured in the CC mice revealed significant correlations between the pink, yellow, salmon, tan, red, and magenta modules with body fat %. Scale indicates the strength of correlations. Correlation values in C are shown in Supplementary Table 10, Additional file 2