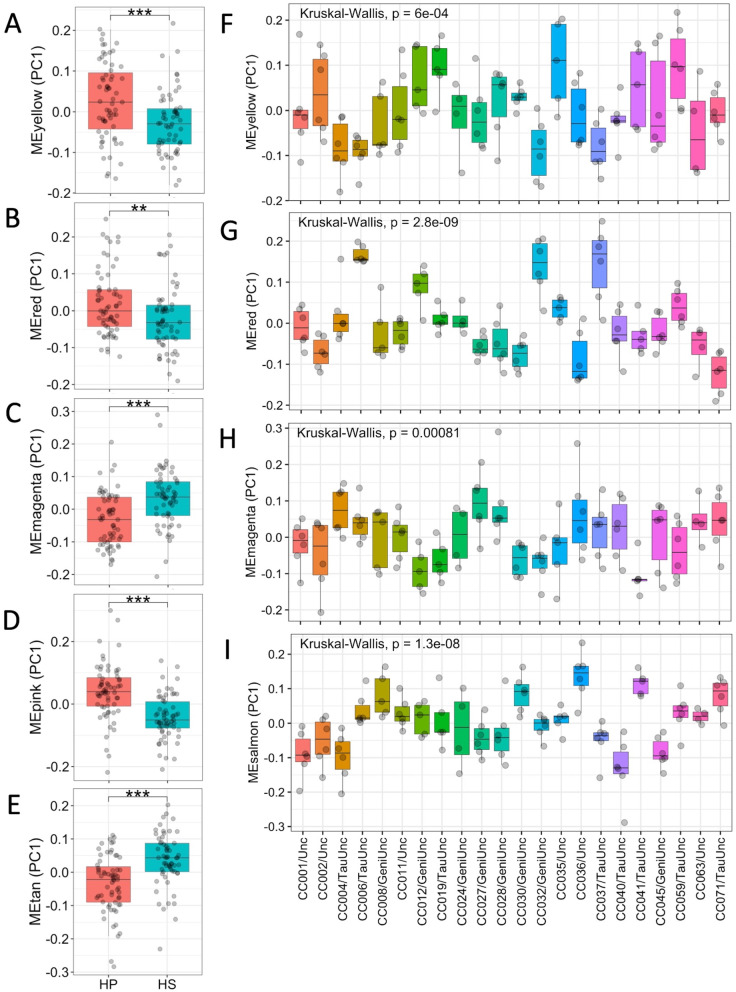
Fig. 7.
Differences in diet macronutrient composition and genetic background perturb MEs (PC1). Most module eigengene (ME) average gene expression profiles (PC1) significantly correlated with body fat % also significantly differed by diet to different degrees, as ascertained with Wilcoxon ranked-sum tests. The MEs that significantly differed by diet were A yellow (p < 0.001), B red (p < 0.01), C magenta (p < 0.001), D pink (p < 0.001), and E tan (p < 0.001), but not salmon (p > 0.1). Most module eigengene (ME) average gene expression profiles (PC1) significantly correlated with body fat % also significantly differed by CC strain to different degrees, as ascertained with Kruskal-Wallis tests. The pink and tan MEs did not differ significantly by CC strain (p > 0.07), but the MEs for the F yellow (p = 6.0 × 10−4), G red (p = 2.8 × 10−9), H magenta (p = 8.1 × 10−4), and I salmon (p = 1.3 × 10−8) modules differed significantly by CC strain. Points indicate individual calculated ME expression for each mouse, and CC strains are ordered numerically