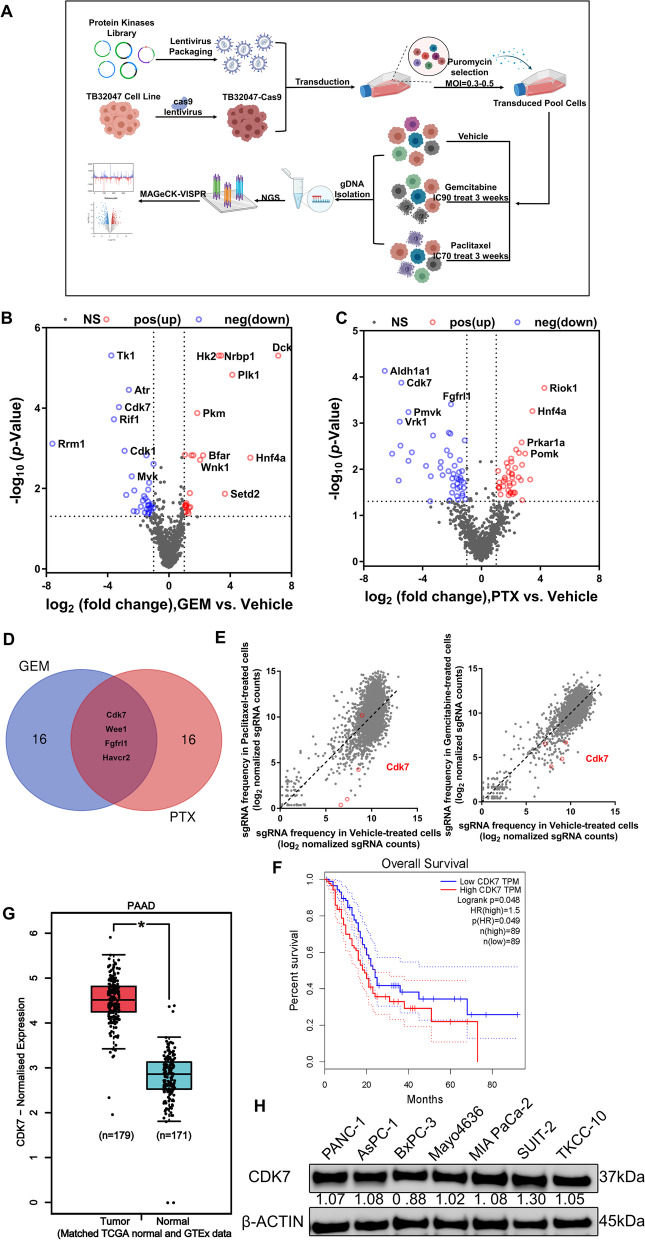
Fig. 1.
CRISPR-Cas9 knockout screens to identify chemosensitivity targets in pancreatic cancer cells treated with gemcitabine and paclitaxel. A Schematic diagram of the timeline and experimental procedure for CRISPR-Cas9 screening using protein kinase library. B, C Volcano plots showing log2-transformed sgRNA normalized fold change and -log10-transformed p-value of gemcitabine (B) and paclitaxel (C) versus vehicle-treated TB32047 cells, with significant genes in positive and negative selection highlighted in red and blue, respectively. D Venn diagram showing the shared genes of the top 20 candidate genes for gemcitabine and paclitaxel negative screening, which are Cdk7, Wee1, Fgfrl1, and Havcr2. E Scatterplots showing log2-transformed sgRNA normalized read counts of gemcitabine (left) and paclitaxel (right) versus vehicle-treated TB32047 cells, with sgRNAs targeting Cdk7 highlighted. F Data from TCGA-PDAC were applied for survival analysis. Kaplan–Meier survival analysis shows that higher CDK7 mRNA expression is associated with a tendency toward poor overall survival. G The analysis of CDK7 expression in PDAC tissues and normal pancreatic tissues was conducted using the TCGA (PDAC) and GTEx (Pancreas) databases. Taken together, the results indicate that CDK7 expression levels were significantly higher in pancreatic cancer than in the corresponding normal pancreatic tissue. H Western blot analysis revealed a variation in CDK7 expression between pancreatic cancer subtypes. * p < 0.05; *** p < 0.001, and **** p < 0.0001 were calculated using the unpaired Student’s t test