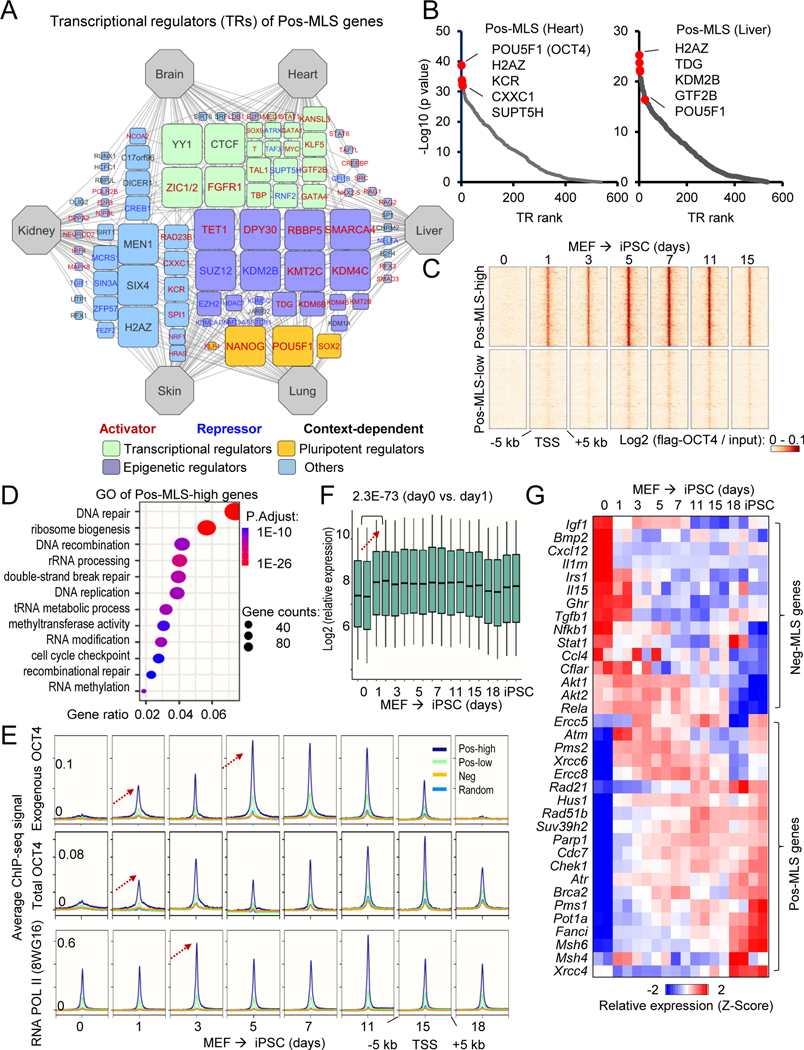
Figure 6. Pos-MLS genes are targets of master pluripotency regulators and are upregulated during somatic cell reprogramming.
(A) Regulatory network visualizing potential TRs of Pos-MLS genes. The connection between tissue and TR indicates that TR shows a significantly higher binding preference to Pos-MLS genes identified in this tissue compared to random genes. The node size (count) indicates the number of tissues in which the genes were identified as top-ranked (30 with lowest p values) regulators. TRs were classified into different functional categories highlighted with different colors. TRs positively or negatively regulating gene expression are shown in red and blue font, respectively. See Methods for details.
(B) Scatter plots showing the TR rank in the heart (left) and liver (right). Y-axis indicates the p values that are calculated using the Wilcoxon rank test comparison of the pos-MLS genes and background. Top regulators with the lowest p values are labeled in red.
(C) Heatmaps showing differential binding activities of pluripotent regulator OCT4 at Pos-MLS genes during somatic reprogramming. Two clusters of genes, Pos-MLS-low (2125 genes) and Pos-MLS-high (1395 genes), were identified by kmeans method according to the low and high binding affinity of OCT4.
(D) The dot plot showing the GO enrichment of genes in the Pos-MLS-high cluster. The colors indicate the BH-adjusted p values. The dot sizes indicate the number of genes.
(E) Metagene analysis showing dynamic binding activities of pluripotent regulator OCT4 and RNA Pol II during the OSKM-induced somatic reprogramming. ChIP-seq data are from (Chen et al., 2016). Arrows indicate the direction of change in ChIP-seq signal.
(F) Boxplot showing the upregulation of Pos-MLS-high genes during the OSKM-induced somatic cell reprogramming. p values were calculated using two-tailed Student’s t-test. Microarray data are from (Chen et al., 2016).
(G) Heatmap showing the expression change of representative Pos- and Neg-MLS genes during the OSKM-induced somatic reprogramming. Each row represents a gene and its expression levels at different times during reprogramming were normalized by Z-score. Microarray data are from (Chen et al., 2016).