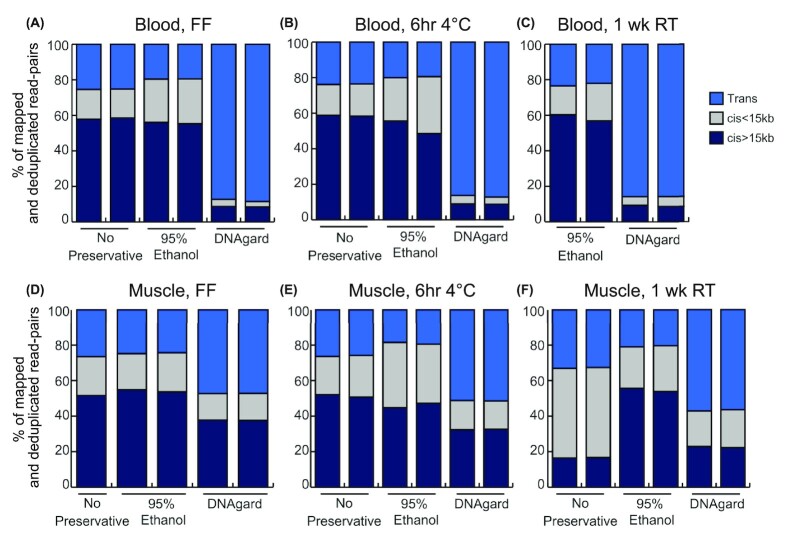
Figure 4:
Hi-C platform benchmarking of bird samples. Stacked bar plots denoting proportions of Hi-C reads mapped to the zebra finch reference genome involving different chromosomes (trans), on the same chromosome but less than 15 kb apart (cis <15 kb), and on the same chromosome and greater than 15 kb apart (cis >15 kb). Tested samples include blood samples (A–C) and muscle samples (D–F). The desirable outcome is to have much greater proportions of Hi-C reads being long-range cis pairs, which reflects an efficient capture of long-range interactions needed for genome scaffolding and haplotype phasing. Hi-C data were generated by Arima Genomics following their standard protocol.