Abstract
Background
The characterization of reinfection with SARS-CoV-2 has been a subject of concern and controversy, especially with the surge of infections with highly transmissible variants worldwide.
Methods
This retrospective national study used comorbidities, vaccination status, SARS-CoV-2 variants of concern, and demographics data to profile participants who were reinfected with SARS-CoV-2, defined as having two reverse transcriptase-polymerase chain reaction-positive SARS-CoV-2 tests within at least 90 days apart. A multivariate logistic regression model assessed the risk factors associated with reinfection . Two control groups were selected: nonreinfected participants reporting a positive test (control group one) and those reporting a negative test (control group two).
Results
Between March 2020 and December 2021, 4454 reinfected participants were identified in Saudi Arabia (0.8%, 95% confidence interval [CI] 0.7-0.8). The majority (67.3%) were unvaccinated (95% CI 65.9-68.7) and 0.8% (95% CI 0.6-1.1) had severe or fatal SARS-CoV-2 disease. COVID-19 vaccines were 100% effective against mortality in reinfected individuals who received at least one dose, whereas it conferred 61% (odds ratio [OR] 0.4, 95% CI 0.1-1.0) additional protection against severe disease after the first dose and 100% after the second dose. In the risk factor analysis, reinfection was highly associated with comorbidities, such as HIV (OR 2.5, 95% CI 1.3-5.2; P = 0.009), obesity (OR 2.3, 95% CI 1.3-3.9; P = 0.003), pregnancy (OR 3.2, 95% CI 1.4-7.4; P = 0.005), and working in health care facilities (OR 6.1, 95% CI 3.1-12.9; P <0.0001). The delta variant (B.1.617.2) was the most frequent variant of concern among the reinfected cohort.
Conclusion
This in-depth study of the reinfection profile identified risk factors and highlighted the associated SARS-CoV-2 variants. Results showed that naturally acquired immunity to SARS-CoV-2 through multiple reinfections together with vaccine-induced immunity provided substantial protection against severe SARS-CoV-2 disease and mortality.
Keywords: SARS-CoV-2 Reinfection, Vaccination, Protective Immunity, Variant Of Concern, Risk Factor, Saudi Arabia
Introduction
The emergence of SARS-CoV-2, a virus that causes COVID-19, has infected over 430 million people worldwide (743,205 in Saudi Arabia) and resulted in over 6 million deaths (8993 in Saudi Arabia) as of February 25, 2022. Despite implementing stringent control measures and travel restrictions, COVID-19 continues to circulate globally, and more recently, the resurgence of COVID-19 cases has been observed after the relaxation of lockdown and social distancing procedures as well as the emergence of variants that posed an increased risk to global public health (Tillett et al., 2021).
It was on August 25, 2020 that the first case of reinfection after 4.5 months of initial infection was reported (To et al., 2021), which was subsequently followed by additional cases, each with variable symptom severity on reinfection, from India, the Netherlands, Belgium, and Ecuador (Gupta et al., 2021; Prado-Vivar et al., 2020; Van Elslande et al., 2021). These studies have shown that the viral genome in primary infections and secondary reinfections are phylogenetically distinct and belong to different evolutionary lineages, suggesting that the virus strain detected in the second episode is very different from the strain found in the first episode.
As the reinfection rate is likely to be a threat to the end of the COVID-19 pandemic, understanding the frequency, duration of protective immunity, clinical impact, and severity of COVID-19 reinfections is essential to predict the course of the pandemic and preparing for future waves. It would also help gain important insights into the pathophysiology of COVID-19 and guide ongoing vaccine development efforts and future public health surveillance policies. Moreover, predicting who is susceptible to being reinfected is critical for risk evaluation assessments (Babiker et al., 2021).
Given the low supply of vaccines at the beginning of the vaccination scheme, one common strategy adopted in many countries (e.g., in France, Germany, Italy, and Spain) was to provide only one dose of vaccine to previously infected individuals. This approach has been supported by several studies showing that a single SARS-CoV-2 infection enables one to acquire natural immunity (Ebinger et al., 2021; Goel et al., 2021; Wang et al., 2021). After the successful implementation of BNT162b2 (Pfizer-BioNTech), ChAdOx1/AZD1222 (Oxford/AstraZeneca), and mRNA-1273 (Moderna) vaccination schemas in Saudi Arabia, with over 60 million receiving their first vaccine dose and 23.6 million vaccinated with two doses, individuals who have been infected are strongly advised by Weqaya, the Saudi center for disease control and prevention (CDC) (2021), to delay their vaccination until 90 days after infection. This strategy relies on the notion that an initial SARS-CoV-2 infection provides powerful protection so that only a single dose of vaccine would be sufficient in those cases. Nevertheless, it is debatable whether a previous SARS-CoV-2 infection or receiving a vaccination can achieve adequate immunity against symptomatic reinfection (Jain et al., 2021).
Moreover, the surge of delta (B.1.617.2) and, more recently, omicron (B.1.1.529) variants has raised major concerns regarding the potential decline in vaccine effectiveness (Lopez Bernal et al., 2021; Collie et al., 2022). Largely dominant worldwide, the delta variant has been proven to have increased replication, leading to higher viral loads and subsequently greater transmissibility than its alpha and beta counterparts (Planas et al., 2021; Sheikh et al., 2021; Vasireddy et al., 2021). Little is known about the sensitivity of emerging variants of concern to the humoral immune response; therefore, individuals who were reinfected with SARS-CoV-2 and harboring these variants will be studied in more detail.
Herein, we present, to the best of our knowledge, the first study in Saudi Arabia to report a comprehensive demographical, clinical, and molecular profiling of SARS-CoV-2 multi-reinfected cases in a mass testing and vaccination setting.
Methods
Study Population, Design, and Data Collection
A total of 4454 reinfected individuals were included in this study over the period from March 1, 2020 to December 1, 2021, constituting the longest national follow-up period. A duration of at least 90 days between two consecutive positive reverse transcription-polymerase chain reaction (RT-PCR) test results for SARS-CoV-2 was the criteria used to identify cases with a high risk of suspicion for SARS-CoV-2 reinfections, following guidelines from the United States CDC (CDC, 2020a, 2020b). Remarkably, although the majority of individuals (4258) have acquired at least two episodes of SARS-CoV-2 infection within at least a period of 90 days (denoted as x1 reinfection), very few acquired multiple reinfection episodes, with 196 individuals having suspected three infections (x2 reinfections).
Reinfected patients’ data were extracted electronically from the centralized Saudi ministry of health (MOH) data repositories, including all SARS-CoV-2 RT-PCR results. Precisely, clinical and demographic characteristics, vaccination status, variant sequencing data, severe disease defined as patients admitted into intensive care unit (ICU), and mortality (with a follow-up until January 30, 2022) were derived for all reinfected cases. Self-reported data obtained from the vaccination program were extracted to learn about the clinical and social characteristics of the cohort (e.g., comorbidities, occupation). This retrospective study was approved by the institutional review board from the Saudi MOH (IRB Log No. 21-111 M), with a waiver of written informed consent.
SARS-CoV-2 Laboratory Testing and Viral Genome Sequencing
Viral genome sequencing was conducted to determine the variants of concern associated with reinfections across a subset of SARS-CoV-2 cases, subject to sample availability and whenever it was possible to retrieve both the primary and reinfection swabs. Viral specimens with cycle threshold values of ≤30 were selected for sequencing by the public health laboratory (Saudi CDC). Briefly, an amplicon-based enrichment approach was used for whole-genome sequencing, using the ATOPlex SARS-CoV-2 full-length genome panel, according to the manufacturer's instructions. Paired-end sequencing was performed on the DNBSEQ-G400 MGI platform. Paired reads were trimmed and mapped to SARS-CoV-2 reference genome sequence MN908947.3 using Burrows-Wheeler Aligner (version 0.7.17), followed by variant calling by SAMtools Mpileup and BCFtools. Consensus sequences were generated by BCFtools and assigned to SARS-CoV-2 lineages with Pangolin.
Statistical Analyses
Clinical and demographic characteristics of the reinfected cohort were described with frequency distributions and measures of central tendency. Vaccination status was compared between vaccinated and unvaccinated reinfected individuals based on the number of doses, the type of vaccine administered, and the disease severity (intensive care hospitalizations and/or mortality). Risk factor analysis was conducted by investigating the profile of reinfected, primarily infected, and uninfected individuals. Cases are reinfected individuals denoted as x1 reinfection, whereas controls are nonreinfected individuals with or without at least one positive RT-PCR (n = 4454 for both control groups), selected from May 20, 2021 to December 01, 2021, to reduce the bias caused by reopening of the country with the surge of variants and fewer restrictions. The first control group was therefore considered participants “at-risk” of reinfection, whereas the second control group distinguished reinfected from uninfected individuals. The odds ratios (ORs) for 20 risk factors associated with reinfection, including age, sex, comorbidities, working in health care, health conditions, and vaccination status, were retrieved using a multivariate logistic regression model (Supplementary Methods). The accuracy of the model was evaluated using the area under the curve against the testing dataset corresponding to all the x2 reinfections cohort (therefore independent of the training dataset, n = 196), with a control group (n = 196) comprising primary infected individuals. The model should predict 1 if cases are PCR-positive and 0 if cases are PCR-negative. Statistical significance was determined at a P-value of <0.05. The following statistical analyses tools were used in this study: Python 3.7 with the packages NumPy (version 1.19.2), Pandas (version 1.1.3), SciPy (version 1.5.2), and statsmodels (version 0.12.1); R with the packages glmnet (version 4.1.1), pROC (version 1.18.0), and forestplot (version 2.0.1).
Results
Demographic and Clinical Features of SARS-CoV-2 Multi-Reinfected Cases
To elucidate the frequency and characteristics of SARS-CoV-2 reinfections among our cohort, 4454 COVID-19 reinfected cases were collected retrospectively; of this, 63.7% (n = 2712) of x1 reinfected and 68.9% (n = 135) of x2 reinfected individuals were male and surprisingly young, falling in the (26-35 year) age group, with the median age of onset being 32 and 29 years old, respectively, as presented in Table 1 . The time lapses between each consecutive SARS-CoV-2 infection episode varied considerably, ranging from 90-600 days. In our cohort, over 60% of x1 reinfected cases (n = 2819) had longer disease intervals, between 200 and 600 days (Table 1). Notably, of the whole cohort, approximately 22% (n = 960) of variant sequencing data were collected, as shown in Table 1, corresponding to those who arrived from overseas as a result of reopening the Saudi borders in May 2021. The majority (88.2%; n = 847) of these reinfected individuals harbored delta (B.1.617.2) variants and delta plus (AY.1) with K417N mutations, followed by beta variants (5.1%; n = 49). Indeed, delta variants started spreading profusely and uncontrollably from July 2021 onwards and could potentially explain the sudden surge in COVID-19 cases in Saudi and reinfection frequency over the summer of 2021.
Table 1.
Demographic and clinical characteristics of individuals who were multi-reinfected with COVID-19.
| Characteristic | No. (%) | |
|---|---|---|
| x1 Reinfected Individuals | x2 Reinfected Individuals | |
| Total | 4258 | 196 |
| Sex | ||
| Female | 1546 (36.3) | 61 (31.1) |
| Male | 2712 (63.7) | 135 (68.9) |
| Age, years | ||
| Median | 32 (IQR [24-40]) | 29 (IQR [25-39]) |
| Mean | 34 (SD 13) | 33 (SD 12) |
| 13-25 | 1098 (25.8) | 53 (27.0) |
| 26-35 | 1696 (39.8) | 85 (43.4) |
| 36-45 | 813 (19.1) | 29 (14.8) |
| 46-55 | 357 (8.4) | 13 (6.6) |
| 56-65 | 175 (4.1) | 14 (7.2) |
| ≥66 | 119 (2.8) | 2 (1) |
| Time Lapse Between Infectionsa | ||
| From 90 to 100 days | 187 (4.4) | 41 (10.5) |
| From 100 to 200 days | 1252 (29.4) | 245 (62.5) |
| From 200 to 600 days | 2819 (66.2) | 106 (27) |
| Variants | ||
| Total | 957 (22.5)b | 3 (1.5)c |
| Delta | 844 (88.2) | 3 (100) |
| Beta | 49 (5.1) | 0 |
| Alpha | 29 (3) | 0 |
| Wild-type | 35 (3.7) | 0 |
| Vaccination Status | ||
| Unvaccinatedd | 2823 (66.3) | 175 (89.3) |
| Vaccinated | 1435 (33.7) | 21 (10.7) |
| Occupation | ||
| Health care Workers | 125 (2.9) | 6 (3.1) |
| Non-Health care Workers | 1976 (46.4) | 97 (49.5) |
| Unknown | 2157 (50.7) | 93 (47.4) |
Time intervals between two infections were counted in days. For x1 reinfections, each individual corresponds to a one-time interval. For x2 reinfections, each individual corresponds to two-time intervals.
For x1 reinfections, both infection episodes were sequenced.
For x2 reinfections, only the second infection episode was sequenced.
Our observation of vaccination and the incidence of reinfection showed that although more than 65% of x1 reinfected individuals were unvaccinated (n = 2823), only 2.3% (n = 96) received two doses of vaccination before the reinfection, where about 60% of cases received their first doses from BNT162b2 (Pfizer-BioNTech) and 40% from ChAdOx1 (AstraZeneca) (Supplementary Table 1). Similarly, 62% of second vaccine doses were administered by BNT162b2 (Pfizer-BioNTech), 30% by ChAdOx1 (AstraZeneca), and only 8% by mRNA-1273 (Moderna). The average time interval between vaccination and subsequent infection ranged from 36-73 days (Supplementary Table 1). Remarkably, of the 957 x1 reinfected individuals with variant sequencing data collected, approximately 42.5% (n = 407) were vaccinated, and the majority (89.9%; n = 366) harbored delta variants (Supplementary Table 2).
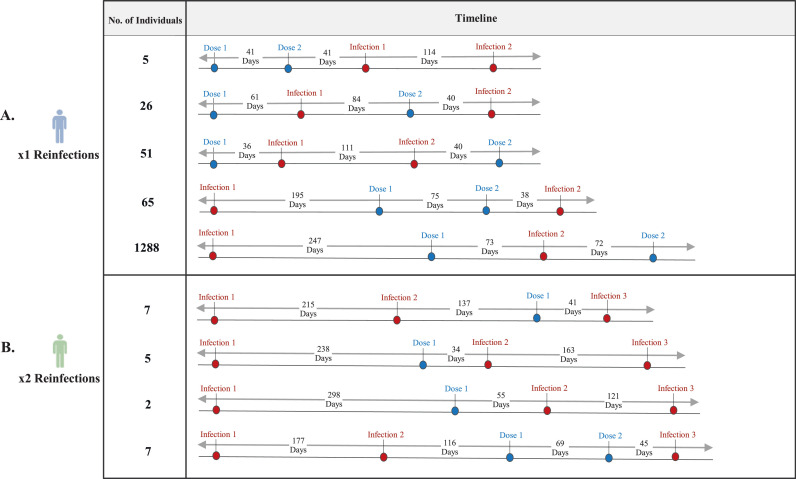
We further investigated all the vaccinated reinfected individuals, grouped based on their SARS-CoV-2 infection and vaccination journey (Figure 1 ). We observed that whilst the minority of x1 reinfected individuals (0.3%; n = 5) developed a breakthrough infection within an average of 41 days (SD 43; median 22 days, interquartile range [IQR] 7-63) after two doses of vaccination and a second infection within an average of 114 days from the first infection; the majority (89.8%; n = 1288) had their primary infections and reinfections flanked by the first vaccine dose, with the time interval between the first vaccination and secondary infection averaged 73 days (SD 41; median 73 days, IQR 37-104) (Figure 1a). Conversely, for the x2 reinfected individuals (n = 196), only 10.7% (n = 21) were vaccinated, with 33.3% (n = 7) of individuals infected for the third time within an average of 45 days after the second vaccine dose (Figure 1b).
Figure 1.
Timeline showing all the different scenarios of SARS-CoV-2 reinfection and vaccination journey.
These scenarios were given across different groups of individuals who were x1 reinfected (a) and x2 reinfected (b) vaccinated, whereby the average time interval between each infection episode (red) and dose of vaccination (blue) is provided in days, regardless of the type of vaccination and severity of symptoms.
COVID-19 Related Comorbidities for Reinfected Cases
We next investigated the association of comorbidities within our cohort to get a better understanding of the population more prone to SARS-CoV-2 reinfection. Among the reinfected cohort studied, we retrieved the comorbidity information from 1029 patients, corresponding to 23.1% of reinfected individuals. It is worth mentioning that a reinfected individual can experience more than one comorbidity simultaneously. Consistent with previous findings that patients who are immunocompromised, such as those who received organ transplants (particularly renal transplants) and patients with uncontrolled diabetes mellitus, are at a higher risk of reinfections with SARS-CoV-2 (Belsky et al., 2021), our data showed that diabetes, followed by hypertension, obesity, and respiratory diseases were the most common comorbidities associated with COVID-19 reinfections among the x1 and x2 reinfected cohort (Table 2 ).
Table 2.
The prevalence of comorbidities associated with COVID-19 reinfections, with confirmed clinical admissions and self-reported data.
| Comorbidities or Immunodeficiencies | No. (%) | |
|---|---|---|
| x1 Reinfected Individuals | x2 Reinfected Individuals | |
| Diabetes | 191 (19.2) | 6 (17.1) |
| Hypertension | 177 (17.8) | 9 (25.7) |
| Obesity | 159 (16) | 7 (20) |
| Respiratory Diseases | 127 (12.8) | 5 (14.2) |
| Cardiovascular Diseases | 116 (11.7) | 2 (5.7) |
| Cancer | 54 (5.4) | 0 |
| HIV Infection | 43 (4.3) | 0 |
| Pregnancy | 42 (4.2) | 1 (2.9) |
| Immunosuppressive Drugs | 24 (2.4) | 1(2.9) |
| Chronic Kidney Disease | 24 (2.4) | 2 (5.7) |
| Sickle Cell Anemia | 22 (2.2) | 1 (2.9) |
| History of Stroke | 8 (0.8) | 0 |
| Organ Transplant | 7 (0.7) | 1 (2.9) |
| Total | 994 | 35 |
HIV, Human Immunodeficiency Virus.
Data are No. (%).
COVID-19 Severe Disease and Mortality Rate for Reinfected Cases
To evaluate the association between SARS-CoV-2 reinfections and severe disease among the x1 reinfected individuals, 31 (0.7%) patients were admitted into the ICU; 26 (83.9%) of these cases were unvaccinated, whereas five (16.1%) had received their first vaccine doses. All these patients received life support during hospitalization, including invasive mechanical ventilation (76.9%), and the median length of hospital stay was 8 days. Similar to non-ICU cases, 50.2% of these x1 reinfected patients in ICU had diabetes, 11.4% had chronic kidney disease, 11% had hypertension, and 6.4% had opportunistic bacterial infections. There were only three (1.5%) ICU cases in the x2 reinfection group, all of whom were unvaccinated, two of them were older than 60 years, and two of them had chronic kidney disease. Furthermore, to determine the frequency of mortality rate due to SARS-CoV-2 reinfections, of the ∼9000 current COVID-19 deaths in Saudi Arabia, four were x1 reinfected cases, all of whom were unvaccinated, and three of them had comorbidities, including hypertension. No death was recorded for the x2 reinfections among our study cohort (Supplementary Table 3). These findings indicate that previous infection confers strong protection against mortality and further reinforces the importance of vaccination to provide immunity against severe COVID-19 and death.
Risk Factors Associated with SARS-CoV-2 Reinfections
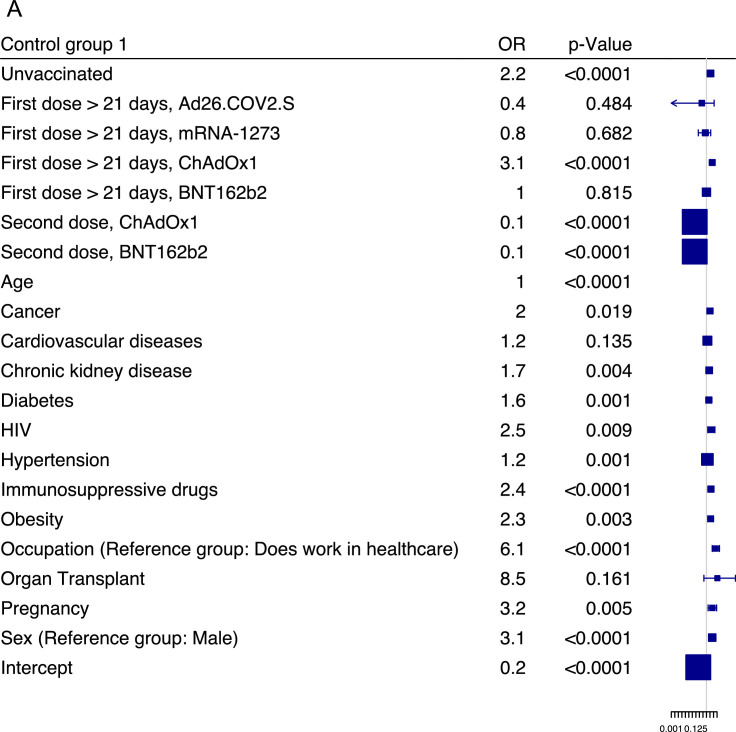
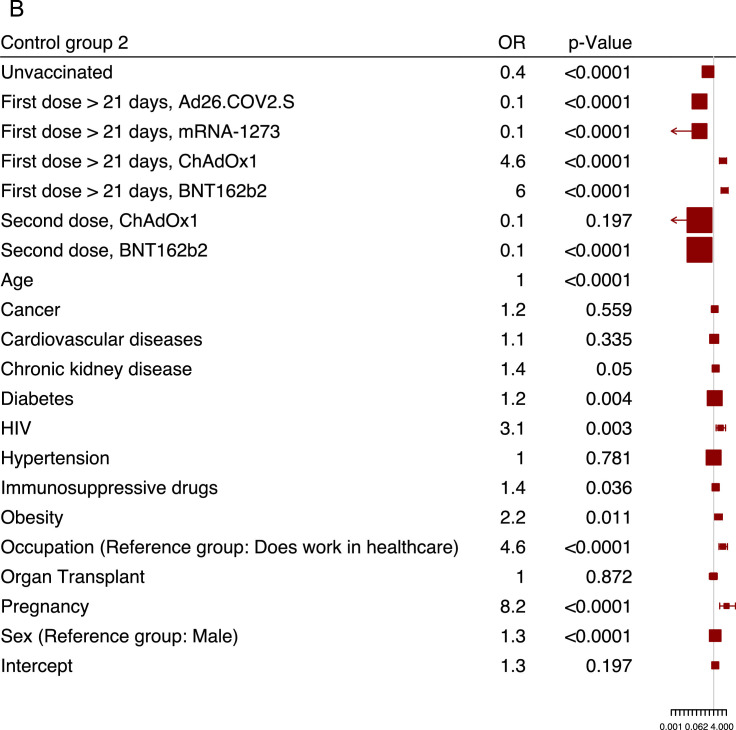
Finally, we determined the risk factors contributing to SARS-CoV-2 reinfections using a multivariate logistic regression model, using x1 reinfection data. The ORs and 95% confidence intervals (CIs) were reported for each risk factor (Supplementary Table 4). The x1 reinfected cases were compared against two control groups: non-reinfected individuals reporting a positive PCR test (control group one) (Figure 2 a) and reporting a negative PCR test (control group two) (Figure 2b).
Figure 2.
Multivariate logistic regresssion analysis of SARS-CoV-2 reinfection risk factors.
Forest plots showing the association between risk factors and SARS-CoV-2 reinfections for a set of demographic and clinical variables for both control groups. The squares and horizontal lines represent the odds ratios (ORs) for each risk factor and their associated 95% confidence intervals (CIs), respectively. An odds ratio larger than one means that the exposure to the risk factor increases the reinfection outcome (and vice versa). (a). Reinfection cases with primarily infected control group one (blue squares). (b). Reinfection cases with noninfected control group two (red squares).
CI, Confidence interval; OR, Odds ratios
As shown in Figure 2a, reinfection was largely associated with vaccination status; unvaccinated individuals were approximately two times more likely to be reinfected than primarily infected individuals (OR 2.2, 95% CI 1.7-2.9; P <0.0001). The probability of those who had their second vaccine doses after 21 days of being reinfected was reduced by more than 87% for both the ChAdOx1 vaccine (OR 0.1, 95% CI 0.1-0.2; P <0.0001) and the BNT162b2 vaccine (OR 0.1, 95% CI 0.1-0.2; P <0.0001) compared with control group one. To determine which comorbidities were associated with a higher risk for reinfections, we observed that HIV infections (OR 2.5, 95% CI 1.3-5.2; P = 0.009), obesity (OR 2.3, 95% CI 1.3-3.9; P = 0.003), followed by chronic kidney disease (OR 1.7, 95% CI 1.2-2.4; P = 0.004) were highly associated with reinfection against primary infected individuals. We observed a similar pattern for control group two, whereby individuals who tested negative were less likely to have the aforementioned comorbidities, except for organ transplant (OR 1.0, 95% CI 0.5-1.8; P = 0.872), although it was not significant owing to the small sample size (Figure 2b). Moreover, individuals known to have a compromised immune system, such as pregnant women and individuals taking immunosuppressive drugs, were more likely to get reinfected, with an average OR of 5.7 and 2.0 for both control groups, respectively. Interestingly, we observed that working in health care was associated with a high risk score of being reinfected (OR 6.1, 95% CI 3.1-12.9; P <0.0001) against control group one and (OR 4.6, 95% CI 2.4-9.6; P <0.0001) against control group two.
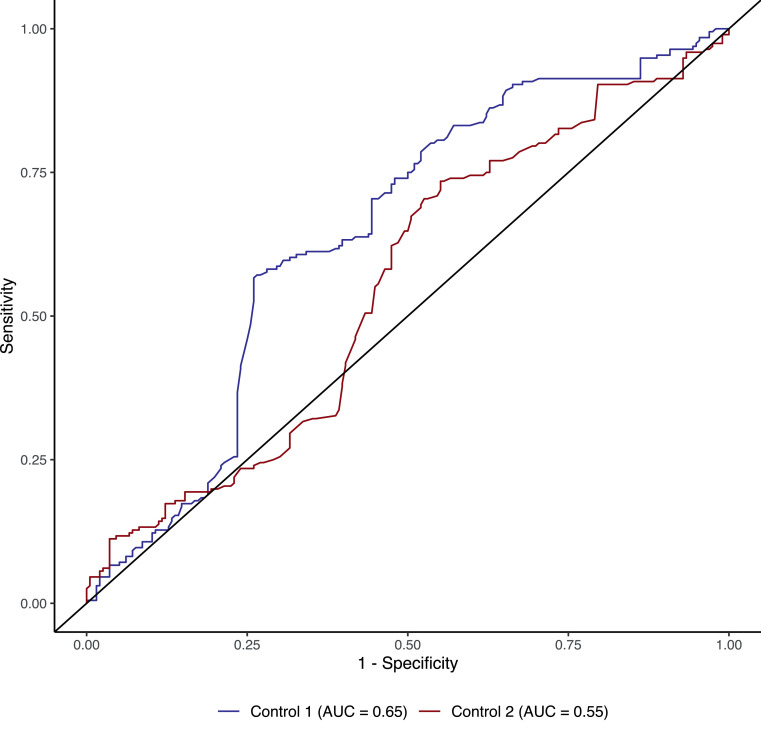
It is worth noting that the ORs for the two control groups were largely consistent throughout, enabling unbiased risk factors analysis and a risk profile for reinfection (Supplementary Table 4). There were, however, some disparities in our analysis. For example, we did not observe any vaccine protection after the first dose for ChAdOx1 and BNT162b2, which suggests a waning in vaccine protection that is consistent with published data (Figure 2b). The predictive power of the multivariate logistic regression model using the x2 reinfection cohort (n = 196) as an independent testing dataset was evaluated to be 65% for control group one and 55% for control group two (Figure 3 ). Such predictive accuracy on new independent data lends stronger support to the results in Figure 2, which depicts several risk factors associated with SARS-CoV-2 reinfection.
Figure 3.
Predictive accuracy of the multivariate logistic regression model.
Prediction model using the x2 reinfection cohort (n = 196) against two nonreinfected control groups; the primarily infected individuals depicted in a blue line (control one) and individuals with negative PCR shown in a red line (control two). The area under the curve (AUC) score is given for each receiver operating characteristic (ROC) curve.
AUC, Area under the curve; ROC, Receiver operating characteristics.
Discussion
To the best of our knowledge, we presented one of the largest cohorts of multiple reinfections with SARS-CoV-2, with insights into their clinical and demographic characteristics and outcomes. It is worth noting that this study was conducted over a 20-month follow-up period (March 01, 2020 to December 01, 2021), encompassing three pandemic waves, with the third wave associated with the highly transmissible delta variant. All the data were extracted retrospectively, with individuals reinfected with SARS-CoV-2 meeting the criteria of more than 90 days between each infection episode regardless of their symptoms, thereby reducing any bias in the selection process.
We reported in this study 4258 x1 reinfections and 196 x2 reinfections, corresponding to a reinfection rate of 0.8% (95% CI 0.7-0.8; P <0.0001) in Saudi Arabia from the period of March 01, 2020 to December 01, 2021. This is akin to an earlier report on reinfections from our neighboring country, Qatar, which estimated the reinfection rate at 0.7 per 10,000 individuals (95% CI 0.6-0.8) (Abu-Raddad et al., 2021d). However, looking at a large observational study conducted among more than 500,000 individuals in Denmark in 2020, 0.7% of individuals who tested positive by PCR in early 2020 tested positive again in late 2020 (Hansen et al., 2021). Nevertheless, none of these studies included a follow-up duration of more than a year, and most studies were completed before the identification of the beta and delta variants (Murchu et al., 2021). In our study, reinfections occurred within an average of 8.4 months (SD 3.2; median 8.9 months; IQR 5.5-11.1) after the first infection episode.
Reinfection suggests that the immune response to a primary infection alone was not adequate to provide sufficient protection against secondary infections. However, we reported that the majority of reinfected individuals were unvaccinated and had comorbidities. This supports previous findings that antibody levels in natural infection are not high enough without vaccination and that the population known to be immune-deficient will be more prone to reinfection, including health care workers (Antonelli et al., 2022; Belsky et al., 2021; Overbaugh, 2020). Therefore, to reduce the likelihood of future reinfections, the study suggests targeting this specific vulnerable group for COVID-19 vaccination, even if they have been previously infected with SARS-CoV-2. This is consistent with many studies, showing the additional protectivity of vaccines against infection among infected populations (Abu-Raddad et al., 2021a, 2021b; Townsend et al., 2021). Surprisingly, our study cohort consisted of young adults, with the majority of reinfected individuals under the age of 30. This could be explained by recent census data from the Saudi general authority for statistics (2021) that more than 70% of the Saudi population are under the age of 30. Overall, we have observed that vaccination reduced the hospitalization and mortality rate among reinfected individuals. This finding supports CDC recommendations that all eligible persons be offered COVID-19 vaccination, regardless of previous SARS-CoV-2 infection status.
Although our sequencing data are limited, we were still able to describe the association between the recrudescence of reinfected cases in Saudi Arabia and the reopening of the Kingdom, coinciding with the emergence of variants, supported by the high prevalence of delta variants in our cohort and the vaccination status among the sequenced subgroup of individuals, demonstrating a waning in vaccine protection against SARS-CoV-2 infection. However, we have shown here that severe illness resulting in intensive care hospitalizations and death in reinfected individuals is a rare phenomenon and mainly occurs among the unvaccinated group, suggesting acquired protection from multiple episodes of infection as shown by several studies (Abu-Raddad et al., 2021a, 2021c). Multiple reinfections and vaccination, so-called hybrid immunity, should provide strong immune protection against severe and fatal diseases, which is reassuring given the frequent resurgence of cases worldwide (Abu-Raddad et al., 2021c; Jeffery-Smith et al., 2021). However, a worrying new variant called omicron (B.1.1.529), first detected in South Africa on November 24, 2021 and later in Saudi Arabia on December 01, 2021, has shown its capability to evade natural and vaccine-induced immunity (Chaguza et al., 2022; Wilhelm et al., 2021). Indeed, reinfections due to omicron have already been reported to be more frequent (Pulliam et al., 2022). Further studies on omicron will be needed, although early reports do not indicate a more severe disease (Karim and Karim, 2021).
Nevertheless, there remains to be further investigations: the risk of reinfection is likely to be related to the antigenic drift of SARS-CoV-2 toward an immunoresistant profile rather than a decline in acquired immunity, as most cases reported in this study were based on a positive RT-PCR test without genomic sequencing to confirm reinfection. Another source of concern is the resurgence of infections among those vaccinated with three doses (Kuhlmann et al., 2022). Further analysis of individuals with multiple reinfections who received a third vaccine dose would be useful to explain the omicron wave observed from December 2021 worldwide.
Acknowledgments
Conflict of Interest Disclosures
The authors have no competing interests to declare.
Data Sharing
The data that support the findings of this study are available at the Saudi MOH, but restrictions apply to the availability of these data, which were used under license for the current study and so are not publicly available. We will consider deidentified participant data sharing upon request after publication.
Funding Source
This work was supported by the Saudi MOH in Riyadh, Saudi Arabia. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Ethical Approval Statement
This study was approved by the institutional review board of the Saudi MOH (IRB Log No. 21-111 M), with a waiver of informed consent.
Acknowledgments
The authors would like to acknowledge their collaborators, Dr. Ahmed Albarrag and Dr. Abdullah Algwizani, from the Saudi CDC/Public Health Authority, for providing variant genome sequencing data
Author Contributions
All authors were responsible for aspects of the study design, data collection, data analysis, and manuscript writing. MAO conceived the study and designed the analysis plan together with MAA, IK, AAS, JA, and MAM. IK performed the data collection and analysis with TA and MHQ. MAO, AAS, IK, and MAM wrote the manuscript. MAO, IK, AAS, JA, MAM, MH, MAA, and EZ contributed to the data interpretation, background literature search, and review. AAG and AAB provided variant sequencing data. All authors read, reviewed, and approved the final submitted manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.07.025.
Appendix. Supplementary materials
References
- Abu-Raddad LJ, Chemaitelly H, Ayoub HH, Tang P, Coyle P, Hasan MR, et al. Effect of vaccination and of prior infection on infectiousness of vaccine breakthrough infections and reinfections. bioRxiv. 2021 [Google Scholar]
- Abu-Raddad LJ, Chemaitelly H, Ayoub HH, Yassine HM, Benslimane FM, Al Khatib HA, et al. Association of prior SARS-CoV-2 infection with risk of breakthrough infection following mRNA vaccination in Qatar. JAMA. 2021;326:1930–1939. doi: 10.1001/jama.2021.19623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad LJ, Chemaitelly H, Bertollini R. National Study Group for COVID-19 Epidemiology Severity of SARS-CoV-2 reinfections as compared with primary infections. N Engl J Med. 2021;385:2487–2489. doi: 10.1056/NEJMc2108120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad LJ, Chemaitelly H, Coyle P, Malek JA, Ahmed AA, Mohamoud YA, et al. SARS-CoV-2 reinfection in a cohort of 43,000 antibody-positive individuals followed for up to 35 weeks. bioRxiv. 2021 [Google Scholar]
- Antonelli M, Penfold RS, Merino J, Sudre CH, Molteni E, Berry S, et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect Dis. 2022;22:43–55. doi: 10.1016/S1473-3099(21)00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiker A, Marvil CE, Waggoner JJ, Collins MH, Piantadosi A. The importance and challenges of identifying SARS-CoV-2 reinfections. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.02769-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky JA, Tullius BP, Lamb MG, Sayegh R, Stanek JR, Auletta JJ. COVID-19 in immunocompromised patients: A systematic review of cancer, hematopoietic cell and solid organ transplant patients. J Infect. 2021;82:329–338. doi: 10.1016/j.jinf.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Common investigation protocol for investigating suspected SARS-CoV-2 reinfection. https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html, 2020 (accessed 26 December 2021).
- Centers for Disease Control and Prevention. Investigative criteria for suspected cases of SARS-CoV-2 reinfection (ICR). https://www.cdc.gov/coronavirus/2019-ncov/php/invest-criteria.html, 2020 (accessed 26 December 2021).
- Chaguza C, Coppi A, Earnest R, Ferguson D, Kerantzas N, Warner F, et al. Rapid emergence of SARS-CoV-2 Omicron variant is associated with an infection advantage over Delta in vaccinated persons. bioRxiv. 2022 doi: 10.1016/j.medj.2022.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie S, Champion J, Moultrie H, Bekker LG, Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinger JE, Fert-Bober J, Printsev I, Wu M, Sun N, Prostko JC, et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med. 2021;27:981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Bhoyar RC, Jain A, Srivastava S, Upadhayay R, Imran M, et al. Asymptomatic reinfection in 2 healthcare workers from India With genetically distinct severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2021;73:e2823–e2825. doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain VK, Iyengar K, Garg R, Vaishya R. Elucidating reasons of COVID-19 re-infection and its management strategies. Diabetes Metab Syndr. 2021;15:1001–1006. doi: 10.1016/j.dsx.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery-Smith A, Rowland TAJ, Patel M, Whitaker H, Iyanger N, Williams SV, et al. Reinfection with new variants of SARS-CoV-2 after natural infection: a prospective observational cohort in 13 care homes in England. Lancet Healthy Longev. 2021;2:e811–e819. doi: 10.1016/S2666-7568(21)00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim SSA, Karim QA, Omicron S-C. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchu O E, Byrne P, Carty PG, De Gascun C, Keogan M, O'Neill M, et al. Quantifying the risk of SARS-CoV-2 reinfection over time. Rev Med Virol. 2021:e2260. doi: 10.1002/rmv.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbaugh J. Understanding protection from SARS-CoV-2 by studying reinfection. Nat Med. 2020;26:1680–1681. doi: 10.1038/s41591-020-1121-z. [DOI] [PubMed] [Google Scholar]
- Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, Marquez S, Gutierrez B, Rojas-Silva P, et al. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN Journal. 2020 [Google Scholar]
- Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudi General Authority for Statistics. Population estimates: population by age groups and gender. https://www.stats.gov.sa/en/43, 2021 (accessed 20 February 2022).
- Saudi Public Health Authority (Weqaya). Interim guidelines for the use of SARS-CoV-2 vaccine. https://covid19.cdc.gov.sa/professionals-health-workers/interim-guidelines-for-the-use-of-sars-cov-2-vaccine/, 2021 (accessed 13 February 2022).
- Sheikh A, McMenamin J, Taylor B, Robertson C. Public Health Scotland and the EAVE II Collaborators. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Hung IF, Ip JD, Chu AW, Chan W-M, Tam AR, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2021:73. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Hassler HB, Wang Z, Miura S, Singh J, Kumar S, et al. The durability of immunity against reinfection by SARS-CoV-2: a comparative evolutionary study. Lancet Microbe. 2021;2:e666–e675. doi: 10.1016/S2666-5247(21)00219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, et al. Symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2021;73:354–356. doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasireddy D, Vanaparthy R, Mohan G, Malayala SV, Atluri P. Review of COVID-19 variants and COVID-19 vaccine efficacy: what the clinician should know? J Clin Med Res. 2021;13:317–325. doi: 10.14740/jocmr4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. MedRxiv. 2021 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.