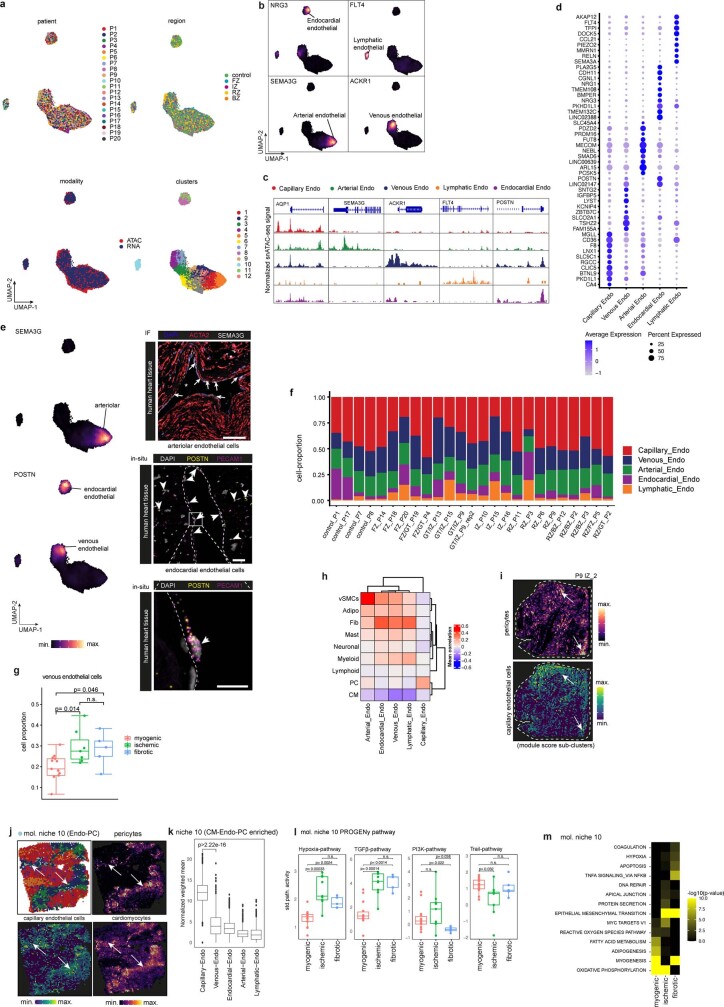
Extended Data Fig. 8. Endothelial cell heterogeneity.
a. UMAP embedding of integrated snRNA-seq and snATAC-seq data coloured by patients, regions, modalities and clusters (resolution= 0.9). b, Gene expression of NRG3, FLT4, SEMA3G and ACKR1. Colours refer to gene-weighted kernel density as estimated by using R package Nebulosa. c, Sub-cluster-specific pseudo bulk ATAC-seq tracks showing the chromatin accessibility of the genes from (b). d, Marker dot plot showing the DEGs for each endothelial cells state and subtype. e, Gene expression of SEM3G and POSTN. Colors refer to gene-weighted kernel density as estimated by using R package Nebulosa. Immunofluorescence of SEMA3G and ACTA2 (upper image). In situ mRNA (RNAscope) for POSTN and PECAM1 (magenta). Arrows highlight endocardial endothelial cells. Scale bars: 75 µm (upper) and 10 µm (lower). f, Endothelial cell state compositions across all patient samples. g, Comparison between patient groups of the venous endothelial cell proportion (two-sided Wilcoxon rank sum test). Each dot represents a patient sample (n for myogenic group = 13, n for ischaemic group = 9, n for fibrotic group = 5). h, Mean Pearson correlation between the abundance of major cell-types and endothelial cell-state scores across all spatial transcriptomics slides. i, Visualization of the spatial distribution of the abundances of pericytes and the state score of capillary endothelial cells sample. Arrows point at colocalization events. j, Visualization of the spatial distribution of molecular niches, the state score of capillary endothelial cells and the abundance of pericytes and cardiomyocytes. Arrows point at locations where molecular niche 10 is present together with high abundances of cardiomyocytes and pericytes. k, Endothelial cell state score distributions in all spots belonging to the molecular niche 10 across all slides (n = 1,874, P-value = 3.19e-298, obtained from a two sided Wilcoxon signed-rank test). l, Mean signalling pathway activities in the molecular niche 10 across different patient groups (adj. P-value of two-sided Wilcoxon Rank Sum test). Each dot represents a slide (n = 14 for myogenic group, n = 9 for ischaemic group, n = 5 for fibrotic group). m, Overrepresented hallmark pathways in the differentially upregulated genes of molecular niche 10 across different patient groups (hypergeometric tests, adj. P-values). In g, k, l data are represented as boxplots where the middle line is the median, the lower and upper hinges correspond to the first and third quartiles, the upper whisker extends from the hinge to the largest value no further than 1.5 × IQR from the hinge (where IQR is the inter-quartile range) and the lower whisker extends from the hinge to the smallest value at most 1.5 × IQR of the hinge, while data beyond the end of the whiskers are outlying points that are plotted individually. In a–c the number of spots of the bottom panels correspond to the barplots in the upper panel.