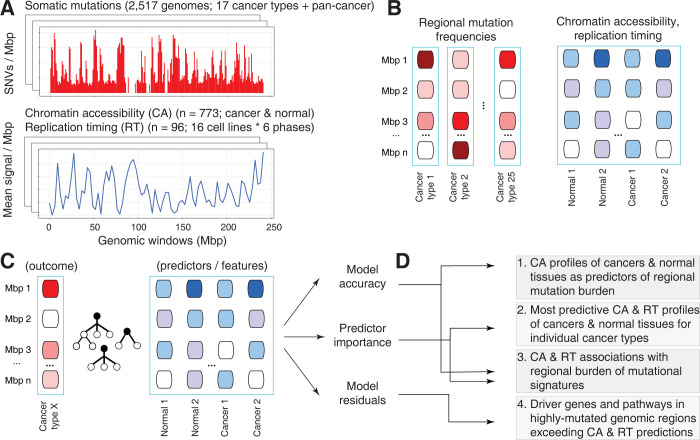
Fig 1. Characterising chromatin accessibility (CA) and replication timing (RT) as predictors of regional mutagenesis in cancer genomes.
A. Somatic mutations in cancer genomes (top) and CA and RT datasets of normal tissues and cancers (bottom) were integrated to study regional mutational processes. Somatic single nucleotide variants (SNVs) of 2,517 whole cancer genomes were analysed with 869 genome-wide epigenetic profiles, including 773 CA profiles of primary human cancers, normal tissues, and cell lines from ATAC-seq experiments, and 96 RT profiles of 16 cell lines and six cell cycle phases from RepliSeq experiments. B. Genomic regions of one megabase (Mbp) were analysed. Regional mutation burden was estimated as the number of SNVs per megabase region. The mean values per region were derived for CA and RT profiles. C. Random forest models were trained using regional mutation burden profiles as the outcome and CA and RT profiles as the predictors. We analysed the pan-cancer dataset and 17 datasets of specific cancer types for which relevant CA and RT profiles were available. D. To associate regional mutation burden with CA and RT, mutational signatures, and cancer driver genes, the random forest models were evaluated in terms of accuracy, predictor importance, and model residuals.