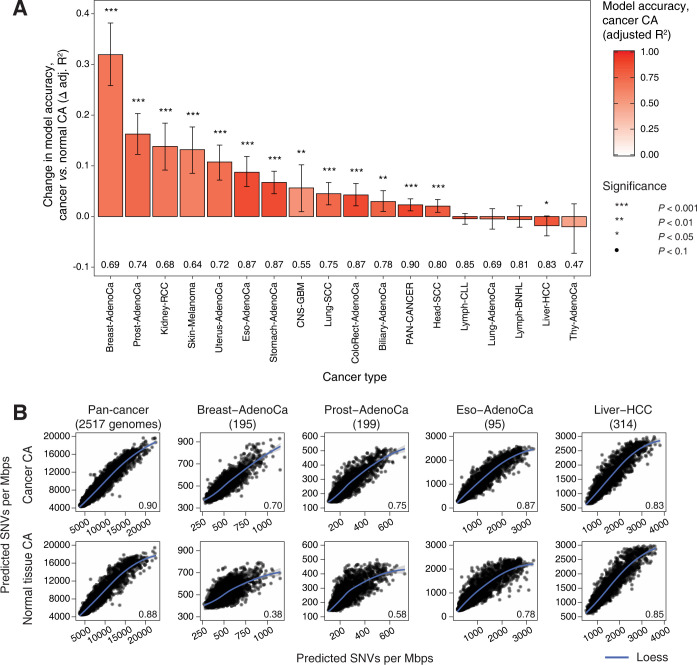
Fig 2. Chromatin accessibility profiles of primary cancers are stronger predictors of regional mutagenesis.
A. Random forest models informed by CA profiles of primary cancers are more accurate predictors of regional mutation burden, compared to models informed by CA of normal tissues. Bar plot shows relative change in prediction accuracy (Δ adjusted R2) of random model regression models informed by CA profiles of primary cancers, compared to matching models informed by CA of normal tissues. Replication timing (RT) profiles are included in all models as reference. P-values of permutation tests and 95% confidence intervals from bootstrap analysis are shown. Accuracy values of models informed by cancer CA profiles are listed below the bars (adjusted R2). B. Examples of regional mutation burden predicted using models informed by CA profiles of cancer (top) vs. CA profiles of normal tissues (bottom). Scatterplots show model-predicted and observed mutation burden (X vs. Y-axis) in one-megabase regions. Prediction accuracy values are shown (bottom right).