Figure 4.
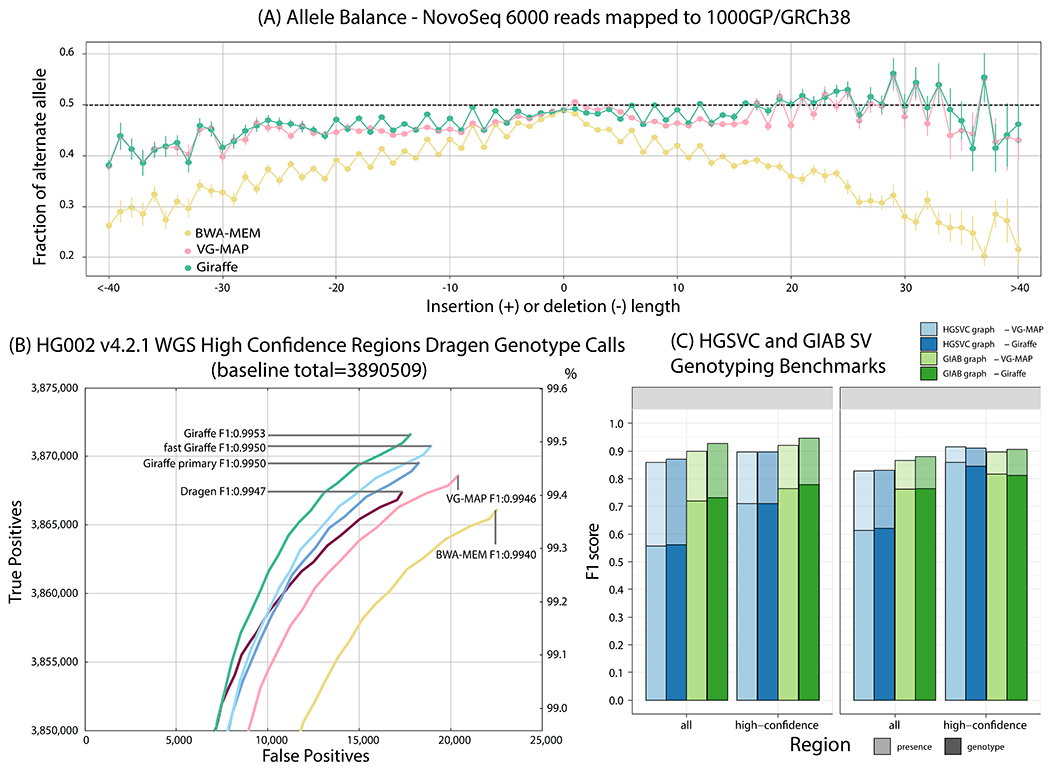
Evaluating Giraffe for genotyping. (A) The fraction of alternate alleles in reads detected for heterozygous variants in NA19239. Reads were mapped to the 1000GP graph with Giraffe and VG-MAP and to GRCh38 with BWA-MEM, and the fraction of reads supporting reference or alternate alleles was found for each indel length. (B) Assessing true positive and false positive genotypes made using the Dragen genotyper with mappings from Giraffe and other mappers. The line labeled Dragen represents the mapper included with the Dragen system itself. (C) Comparing Giraffe to VG-MAP for typing large insertions and deletions. “Presence” (lighter bars) evaluates the detection of SVs without regard to genotype; “genotype” (darker bars) requires the SV to be detected and its genotype to agree with the truth genotype. The y-axis shows the F1 score. For the HGSVC benchmark, we define high-confidence regions as regions not overlapping simple-repeats and segmental duplications. For the Genome in a Bottle consortium (GIAB) benchmark, we use the set high-confidence regions provided by GIAB.
