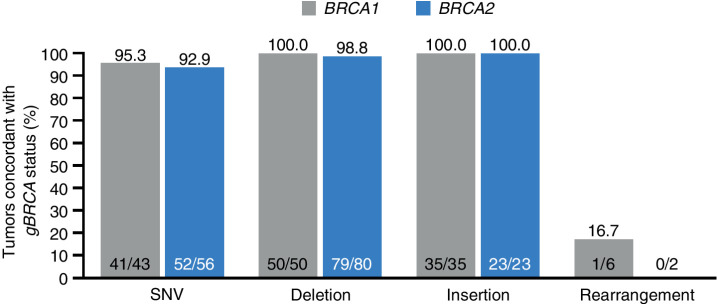
Figure 1.
Tumor sequencing and concordance with germline BRCA status—evaluable ITT population. The proportion of patients with a known gBRCAmut by Central lab who have a BRCA1/2mut (defined as known or likely pathogenic variant, CNAs excluded) detected in a tumor using FoundationOne CDx. All patients showing concordant BRCA1 or BRCA2 mutational status exhibited the same mutation in tumor as originally detected in germline, as evidenced by mapping to a common Variation ID in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) or other comparative means, with one exception (1 patient who exhibited a distinct BRCA2mut in germline and tumor). A total of 14 patients were discordant: 2 patients—same germline variants used to support enrollment were classified by the FoundationOne CDx test as pathogenicity unknown; 7 patients exhibited gBRCA deletion impacting one or multiple exons and were hence mapped to Rearrangement category. These patients exhibited alterations classified as pathogenic BRCA CNAs by FoundationOne CDx in the corresponding BRCA genes; 2 patients—gBRCA2mut, tBRCA1mut; 1 patient—gBRCA1mut, tBRCA2mut; 2 patients—no tBRCA variant detected.