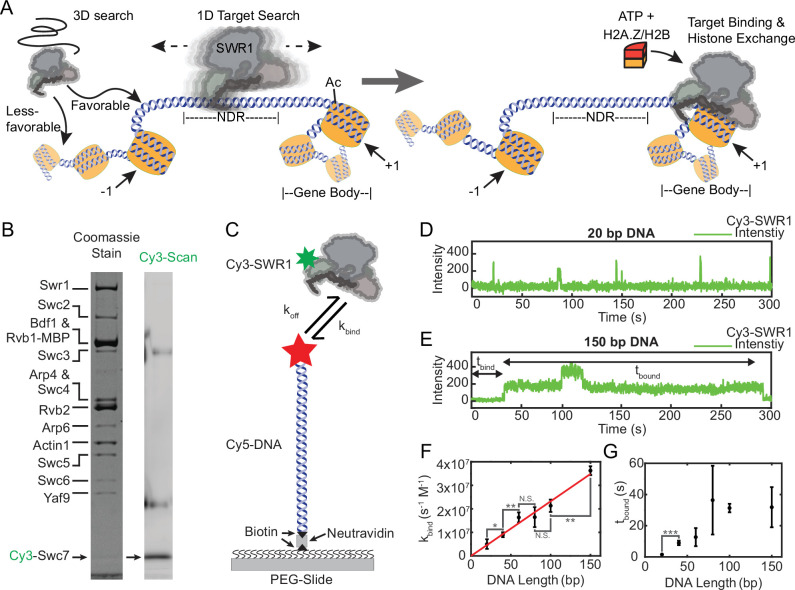
Figure 1. SWR1 binds DNA in short- and long-lived states and prefers longer DNAs.
(A) Proposed facilitated search mechanism for how SWR1 locates the +1 nucleosome. (B) A denaturing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) of reconstituted Cy3-SWR1 imaged for Coomassie (left) and Cy3 fluorescence (right). Cy3-Swc7 is faint when stained with Coomassie but is a prominent band in the Cy3 scan. The two diffuse bands that run at higher molecular weight and appear in the Cy3 scan are carry over from the ladder loaded in the adjacent lane. (C) A schematic of the single-molecule colocalization experiment used for kinetic measurements of Cy3-SWR1 binding to Cy5-labeled DNA of different lengths. Representative traces of Cy3-SWR1 binding to (D) 20 bp Cy5-DNA and to (E) 150 bp Cy5-DNA. A second Cy3-SWR1 can be seen binding at approximately 100 s. (F) The on-rate binding constant for the initial binding event, (kbind) for SWR1 to DNA of different lengths, error bars are standard deviation. N values: 20 bp (40), 40 bp (67), 60 bp (267), 80 bp (129), 100 bp (221), 150 bp (409). The red line is a linear fit to the data, where R2 = 0.99 [two technical replicates represented, statistical differences determine via Student’s t-test, where asterisks indicate the level of significance as conventionally defined (* = p < 0.05; ** = p < 0.01, *** = p < 0.001, n.s. = not significant)]. (G) The lifetime (tbound) of Cy3-SWR1 bound to DNAs of different lengths, error bars are standard deviation. N values: 20 bp (48), 40 bp (118), 60 bp (291), 80 bp (382), 100 bp (339), 150 bp (363) [two technical replicates represented, statistical differences determine via Student’s t-test, where asterisks indicate the level of significance as conventionally defined].