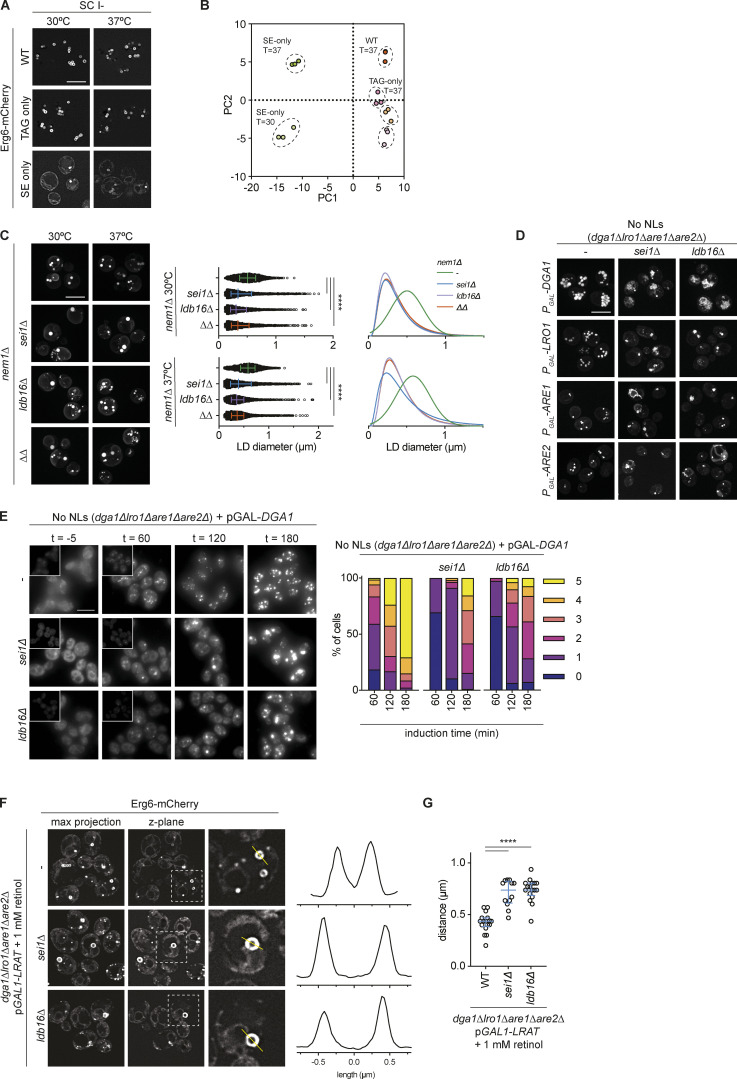
Figure S2.
Sei1/Ldb16 Seipin complex is required for normal morphology and biogenesis of TAG-only and SE-only LDs. Related to Figs. 2 and 3. (A) Fluorescence micrographs of LDs in WT, TAG-only and SE-only cells cultured in inositol free media to late stationary phase at indicated temperature. Erg6 serves as LD marker protein, scale bars correspond to 5 μm. (B) Principal Component Analysis (PCA) of lipid molecular species composition (in mol%) of indicated mutants. Proportion of variance is 58.25% (PC1) and 13.17% (PC2). (C) Analysis of LD morphology in nem1Δ background strains with indicated genotypes. Cells were cultured in inositol free media at 30 or 37°C to late stationary phase, and LDs were stained with BODIPY. Scale bars correspond to 5 μm. (D) Analysis of LD morphology in no NL cells upon plasmid borne overexpression of the indicated NL synthesizing enzyme. Micrographs were taken after 24 h addition of galactose to induce the expression of the NL synthesizing enzymes. LDs were visualized using the NL dye BODIPY 493/503. Scale bars correspond to 5 μm. (E) Analysis of LD biogenesis upon induction of TAG synthesis by expression of the DAG acyltransferase Dga1. Plasmid-borne expression of Dga1 was induced by addition of galactose (final concentration 2%) to cells with the indicated genotype grown in raffinose-containing medium. LDs were stained with BODIPY at indicated timepoints (in minutes after galactose induction). Scale bars correspond to 5 μm. The number of LDs per cell was quantified for a minimum of 50 cells per timepoint. (F) Analysis of LD morphology in cells with indicated genotypes, expressing plasmid-borne human LRAT from the GAL1 promotor. Left: Fluorescence micrographs of Erg6-mCherry (z-max projections and single z-planes) are shown. Dashed boxes indicate location of blow-up images (8 × 8 µm), location of intensity profile is indicated by yellow lines. Right: Line intensity plot of Erg6-mCherry signal. Distance between peak maxima is indicative of LD size. LRAT expression was induced by the addition of galactose (final concentration 2%) and retinol was supplemented at 1 mM (in 1% Tergitol NP-40). LDs were visualized using Erg6-mCherry as a LD marker protein after 8 h of LRAT induction and retinol supplementation. Clustered and supersized LDs typical of sei1∆ and ldb16∆ mutants are indicated by red and yellow arrowheads, respectively. (G) Distance distribution of LD size as determined by Erg6-mCherry line intensity analysis (as in F). Circles show individual measurements, line indicates median (± 95% confidence interval). Difference in distributions were analyzed by non-parametric testing (Kruskal–Wallis test; P < 0.0001), followed by Dunn’s multiple comparison testing (****, P < 0.0001).