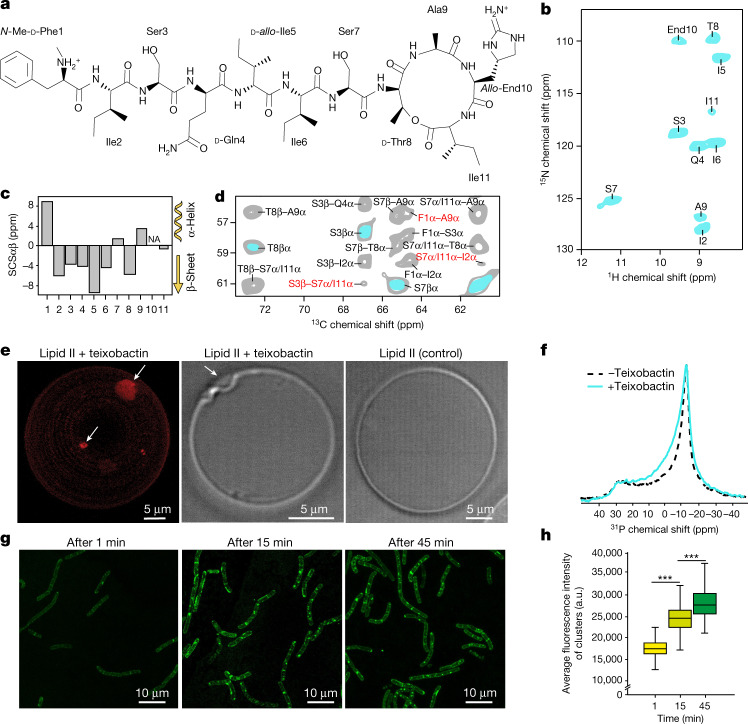
Fig. 1. Oligomerization and membrane remodelling.
a, Chemical structure of teixobactin. b, 2D NH ssNMR spectrum of lipid II-bound teixobactin in membranes. c, Secondary chemical shifts (SCS23) show β-structuring of the N terminus. Source data are provided. NA, not applicable. d, 2D CC ssNMR spectra of lipid II-bound teixobactin acquired with 50 ms (cyan) and 600 ms (grey) magnetization transfer time show intermolecular Cα–Cα contacts (red) consistent with the formation of antiparallel teixobactin β-sheets. e, Visualization of lipid II clustering (arrows) in 3D (left), using confocal microscopy of GUVs doped with Atto-labelled lipid II and treated with teixobactin. The transmission image of the same GUV revealed membrane perturbations induced by the oligomers (middle). Control GUVs with Atto-labelled lipid II, without teixobactin (transmission) (right) are also shown. Note that as the plane used for the middle image is not exactly in the middle of the GUV, the diameter is smaller than for the entire GUV as seen in the left image. The experiment was performed in biological triplicates. f, Static 31P ssNMR spectra show membrane perturbation induced by teixobactin (cyan line) in contrast to the untreated lipid II-doped DOPC large unilamellar vesicles (black dashed line). g, Confocal microscopy with B. megaterium incubated in the presence of a fluorescent teixobactin analogue26. Intense, elongated teixobactin clusters form fast and get more compact after 45 min. h, Boxplot of the fluorescence intensity of clusters after 1, 15 and 45 min of incubation with teixobactin. n = 380 clusters from biological triplicates. Data are represented as a boxplot in which the middle line is the median, the lower and upper hinges correspond to the first and third quartiles, the upper whisker extends from the hinge to the largest value and the lower whisker extends from the hinge to the smallest value, no further than 1.5× interquartile range. ***P < 0.0001, unpaired, two-tailed Student's t-test.