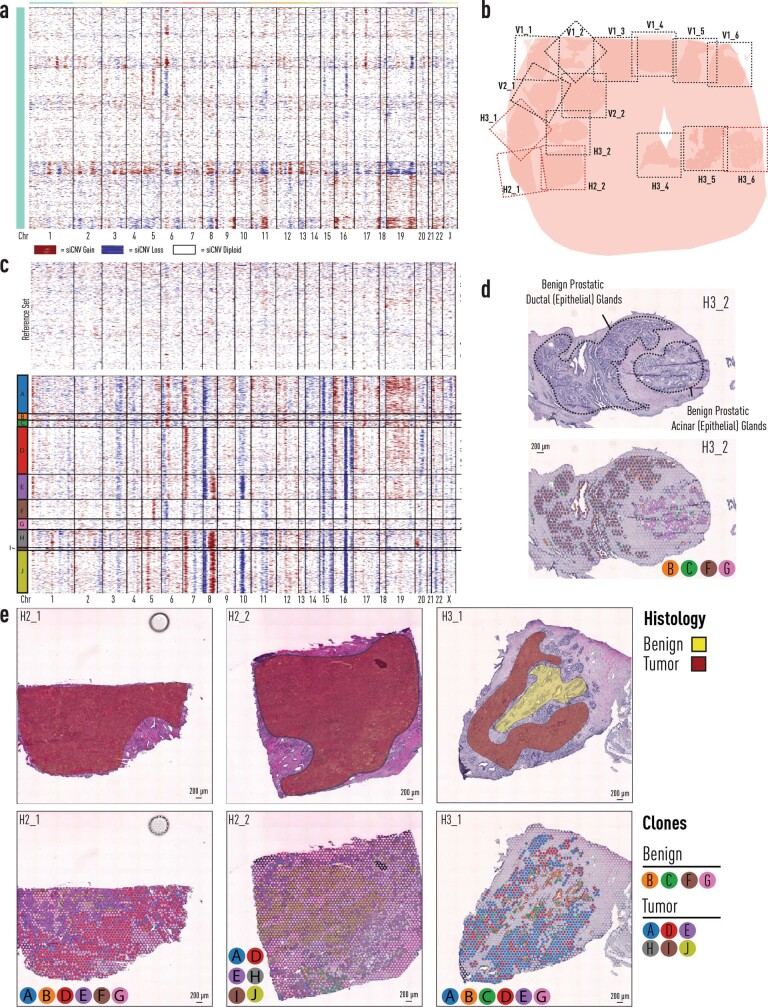
Extended Data Fig. 10. Organscale prostate patient 2.
a, Spatial inferCNV (siCNV) profiles of histologically benign prostatic epithelial cells from 11 sections from prostate patient 2. b, Reference overview of 15 sections available for analysis: sections H2_1, H2_2, H3_1, and H3_6 harbour tumour (marked with red dotted lines). Black dotted lines represent the area covered by spatial transcriptomics array surface. c, Analysis of tumour foci in sections H3_1, H2_1 and H2_2. Analysis includes section H3_2, a non-tumour bearing section which included spatially co-localized benign spots harbouring inferred CNV alterations from panel a. d, Spatial histology and clone distribution in section H3_2 (no-tumour). Benign ductal histology (Clone F) harbours distinct inferred CNVs (chr5 amplification, chr6 deletion), not harboured in neighbouring benign acinar glands (Clone G). e, Section histology (transparent red indicates tumour, and transparent yellow denotes benign) and clones from tumour-bearing sections H3_1, H2_1, and H2_2. CNV = Copy-Number Variant, chr = chromosome.