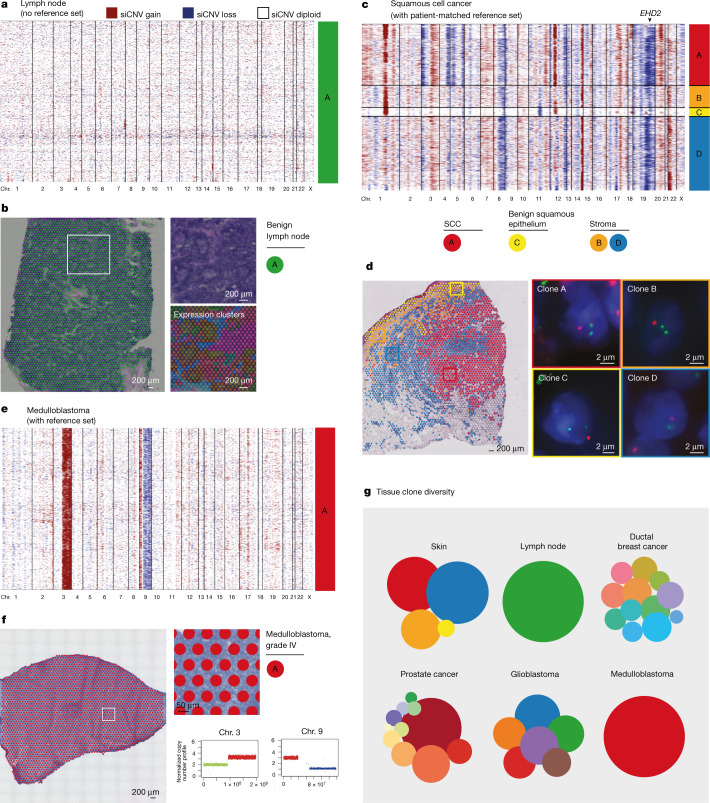
Fig. 4. Somatic copy number alterations in cancer and benign histologies.
a,b, Benign lymph node with distinct histological features and gene expression heterogeneity (b) harbouring no detected copy number alterations (a). Gene expression clusters were determined by UMAP analysis in b. c,d, Skin containing SCC (clone A, red) as well as benign squamous epithelium (clone C, yellow). A subset of somatic events visualized in cancer clone A are also detected in adjacent benign epithelial clone C. d, FISH validation of the siCNV encompassing EHD2 on chromosome 19q (arrowhead in c). e,f, Monoclonal childhood medulloblastoma. siCNVs on chromosomes 3 and 9 (e) were corroborated by copy number calls from WGS (f, lower right). g, Clone distribution for each tissue type. Circle area corresponds to the number of spots per clone. Results from ductal breast cancer and glioblastoma are available in Extended Data Fig. 11.