Figure 3.
3D de-cluster of inactive centromeres delays their replication timing
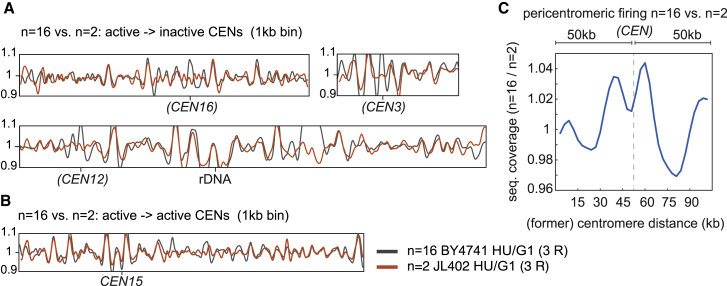
Each replication timing plot41,42 in (A) and (B) is the average representation of 3 independent replicates and shows the sequencing coverage ratio of S phase (HU) synchronized cells normalized on the G1 (α-factor) non-replicating cells (1 kb-binned). Replication timing profiles of the n = 16 (BY4741) are shown in gray, while those of n = 2 size-matched (JL402) are in orange.
(A) Representative comparison profiles of chromosomes with inactive centromeres in n = 2, but active in n = 16 (brackets).
(B) Comparison profiles of chromosome 15 with active centromere in both n = 2 and n = 16.
(C) Pericentromeric firing in n = 16 versus n = 2. The ratio plot shows the early firing of pericentromeric regions (∼100 kb) in n = 16 in respect to n = 2, in which centromeres were inactivated. Centromere position appears in brackets and is indicated with a dotted line.