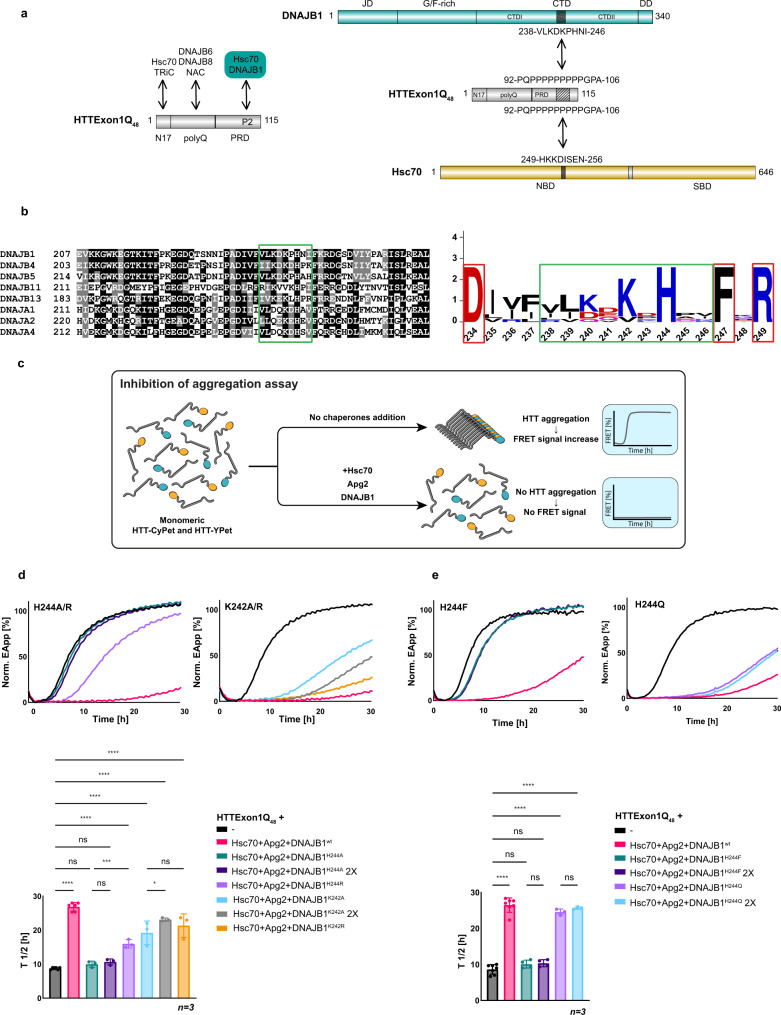
Fig. 1. Identification of a HTTExonQ48-binding motif within DNAJB1 and Hsc70.
a Left, schematic representation of known binding sites of Hsc70, DNAJB1, DNAJB6/DNAJB8, TRiC, and NAC chaperones on HTTExon1Q48. Right, schematic representation of the XL-MS-detected interaction sites between the CTD of DNAJB1 / NBD of Hsc70 and the P2 region of the PRD of HTTExon1. b Left, alignment of human class A and B J-domain proteins (JDPs) containing an amino acid motif similar to the HTT-binding motif (HBM) of DNAJB1 that is framed in green. Right, the sequence logo of the aligned amino acid of the HBM (green frame) shows the positively charged H244 and K242 residues to be most conserved. The flanking amino acids that are also highly conserved are framed in red. c Schematic representation of the FRET-based assay of the analysis of HTTExon1Q48 aggregation. In the absence of chaperones, HTTExon1Q48-CyPet/YPet proteins are in close proximity upon aggregation, which allows energy transfer from excited CyPet to YPet (FRET). The addition of chaperones and ATP prevent HTTExon1Q48-CyPet/YPet aggregation and consequently lead to an absence of FRET. d, e Top, FRET measurements as a readout of HTTExon1Q48 aggregation over time in the absence (black curve) and presence of Hsc70, Apg2 and DNAJB1wt (magenta) or variants (d) H244A, H244R, K242A, K242R, (e) H244F and H244Q. The graphs are representative results of three independent experiments. Bottom, one-way ANOVA analysis of the half-life (T1/2) of HTTExon1Q48 aggregation of the respective analyses using the same color code as shown on top. Bars represent the mean value and error bars correspond to the mean SD. ****P ≤ 0.0001; ***P ≤ 0.001; *P ≤ 0.05; ns not significant.