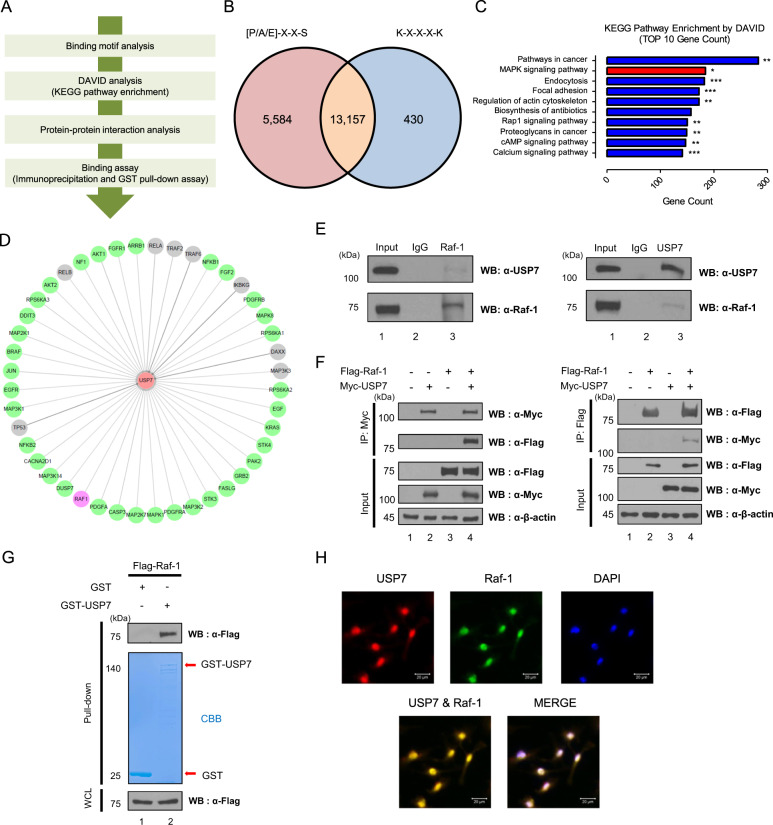
Fig. 1. Motif analysis for identifying new binding partners of USP7.
A Schematic overview of the workflow for identifying new binding partners of USP7 using binding motifs, DAVID, protein-protein interaction, and binding analyses. B The diagram shows the number of proteins having each or both [P/A/E]-X-X-S and K-X-X-X-K motifs. C Pathway enrichment of putative substrate proteins of USP7 was analyzed by DAVID analysis. Y-axis indicates the pathway name, and X-axis indicates gene count in the pathways, and the asterisk indicates the p-value (*p < 0.05, **p < 0.01, ***p < 0.001.) D Interaction network between USP7 and putative substrates with high interaction score and with or without the connection revealed. The color of nodes indicates the confirmation of binding between USP7 and proteins (Gray: published and green: nonpublished), and Raf-1 binding with USP7 is shown in purple. Interaction network is shown by Cytoscape. E Endogenous USP7 or Raf-1 was precipitated with an anti-USP7 or an anti-Raf-1 antibody in HeLa cell lysate. F HEK293T cells were transfected with Myc-USP7, and Flag-Raf-1 and immunoprecipitations were performed by an anti-Myc or an anti-Flag antibody. G Purified GST- or GST-USP7 expressed in BL21 was incubated with Flag-Raf-1-overexpressed HEK293T cells. GST- or GST-USP7 was pull downed by GST bead and gel was stained by Coomassie Brilliant Blue. H Immunocytochemical analysis was performed to investigate the localization of respective USP7 and Raf-1, and co-localization of USP7 and Raf-1.