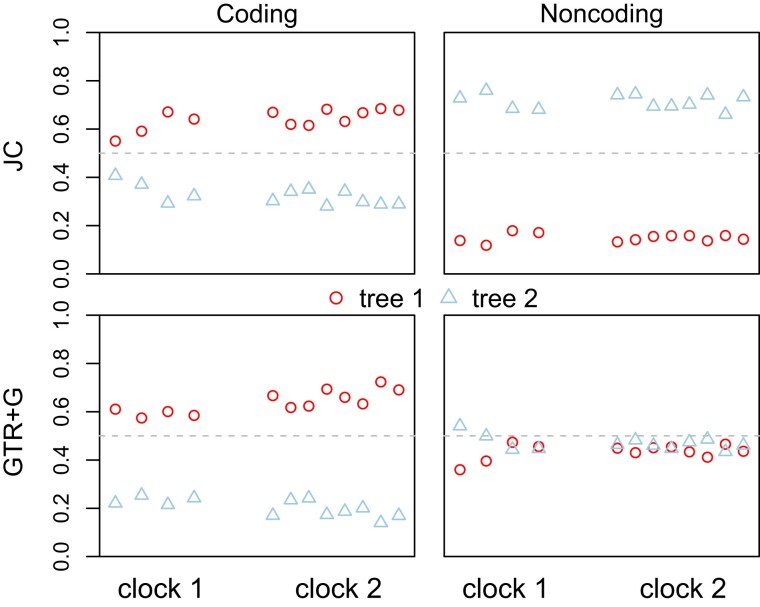
Fig. 8.
Posterior probabilities for species trees 1 and 2 for the gibbons (fig. 7) obtained from bpp analysis of the coding and noncoding datasets under different clock models. In each panel are presented two replicate runs for each of six analyses, specified as (1) clock = 1 (strict clock, one rate for all loci); (2) locusrate = 1 0 0 5 iid, clock = 1 (strict clock, i.i.d. rates among loci); (3) locusrate = 1 0 0 5 iid, clock = 2 10 100 5 iid LN (clock 2, i.i.d. prior for and among loci, and log-normal kernel); (4) locusrate = 1 0 0 5 iid, clock = 2 10 100 5 iid G (clock 2, i.i.d. prior for and among loci, and gamma kernel); (5) locusrate = 1 0 0 5 dir, clock = 2 10 100 5 dir LN (clock 2, dir prior for and among loci, and log-normal kernel); (6) locusrate = 1 0 0 5 dir, clock = 2 10 100 5 dir G (clock 2, dir prior for and among loci, and gamma kernel). The strict clock (clock 1) is assumed in the first two analyses while the independent-rates model (clock 2) is assumed in the next four analyses. The substitution model is either JC or GTR+. Inverse-gamma priors are assigned on and .