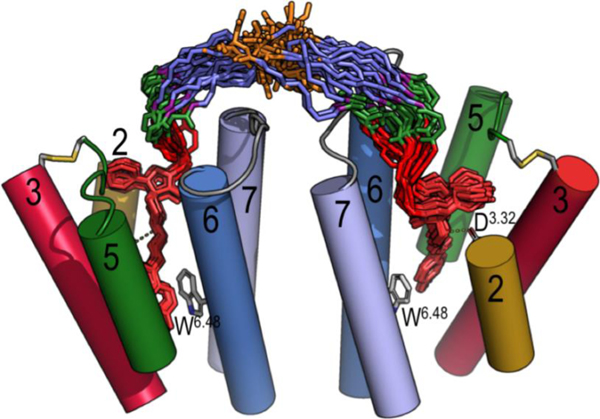
Figure 5.
Evolution of bivalent ligand 13 in the D2R homodimer (TM1 and TM4, ECL1 and part of ECL2 are omitted for clarity), constructed via the TM6 interface, as devised from MD simulations. The structures of 13 (the color code of the atoms is as in Figure 1) are extracted from the simulations (20 structures collected every 50 ns), whereas the structure of the D2R homodimer corresponds to the initial model. A detailed analysis of the simulation (Figure S3) confirms that the designed bivalent ligand 13 remains stable at the orthosteric binding cavities through the unbiased 1 μs MD simulation.