The top five ranked compounds after docking process.
| Metabolite name | Structure | Scoring | RMSD |
|---|---|---|---|
| HA1 |
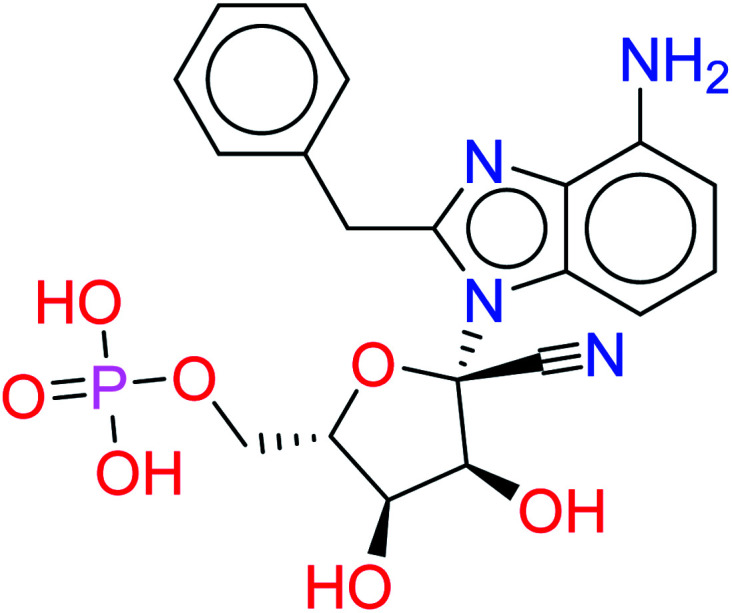
|
−9.7371 | 0.0924 |
| HA2 |
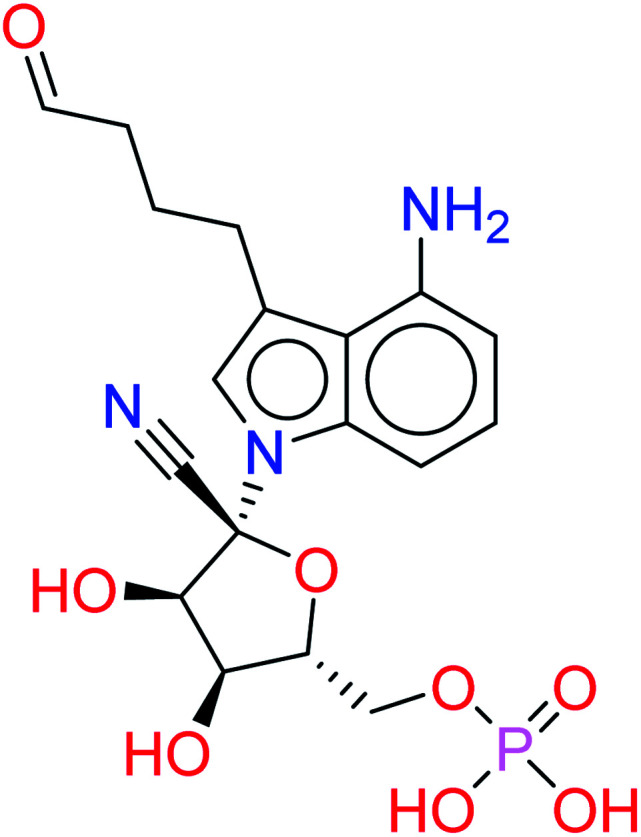
|
−9.6154 | 0.0800 |
| HA3 |
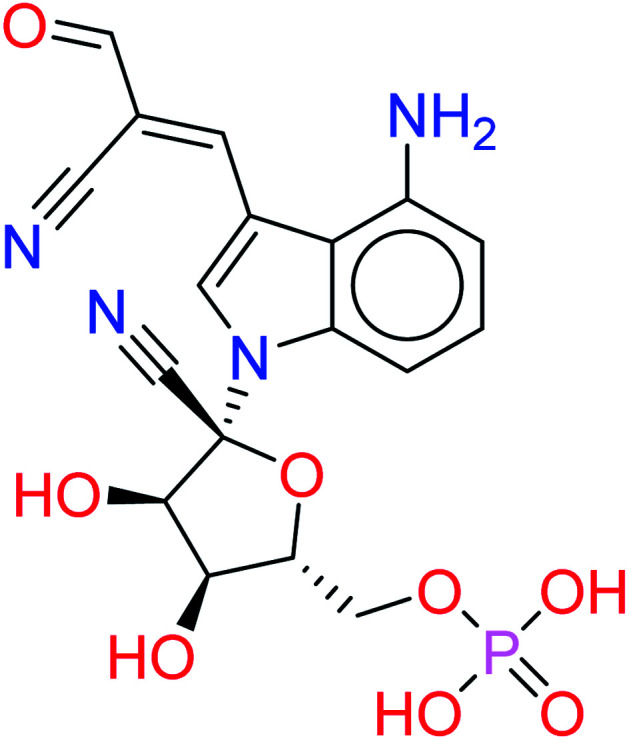
|
−9.4981 | 0.0887 |
| HA4 |
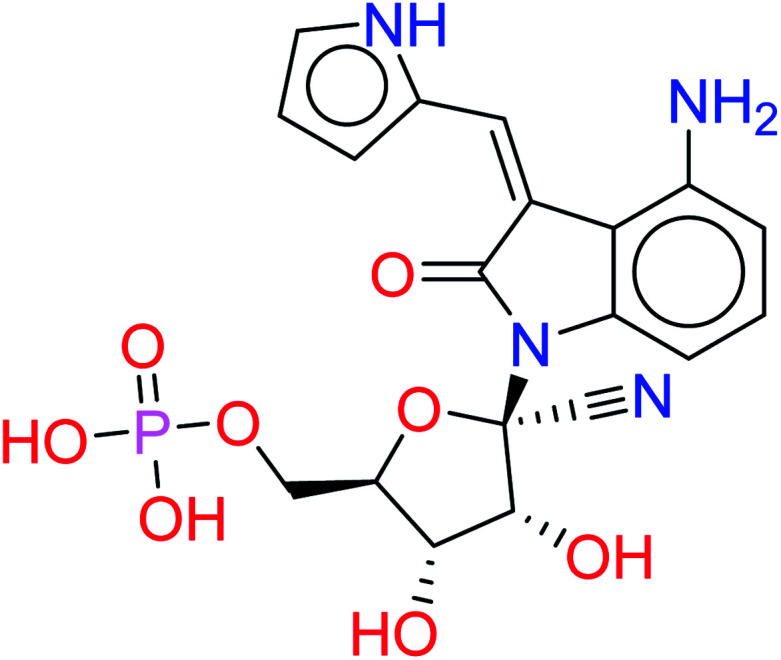
|
−9.3512 | 0.0768 |
| HA5 |
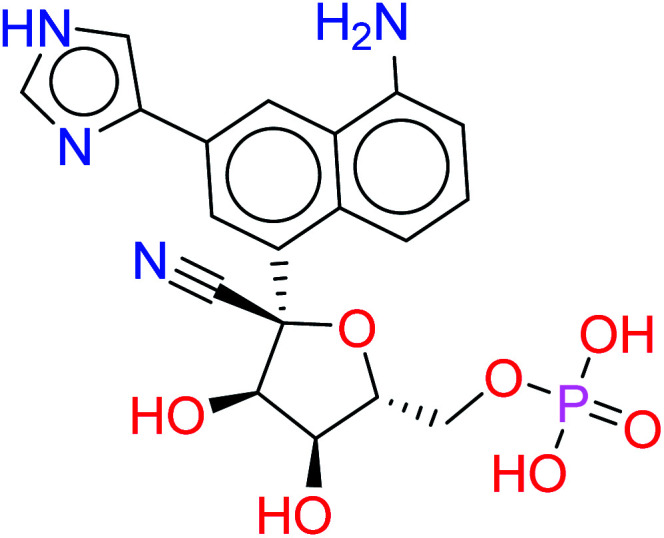
|
−9.2272 | 0.1214 |
| Metabolite name | Structure | Scoring | RMSD |
|---|---|---|---|
| HA1 |

|
−9.7371 | 0.0924 |
| HA2 |

|
−9.6154 | 0.0800 |
| HA3 |

|
−9.4981 | 0.0887 |
| HA4 |

|
−9.3512 | 0.0768 |
| HA5 |

|
−9.2272 | 0.1214 |