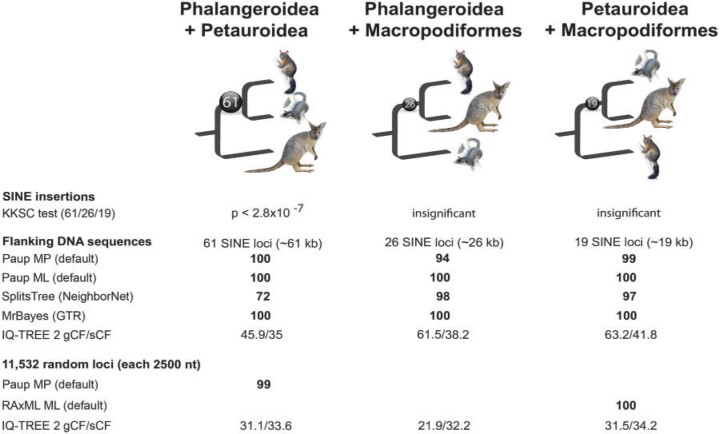
Figure 2.
Compilation of phylogenetic signals for presence/absence SINE markers, their flanking sequence regions, and a random genome sequence data set from 11,532 loci. Sixty-one SINE insertions (black ball) provide significant evidence to merge Phalangeroidea and Petauroidea (KKSC test, 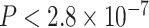 ). Sequence flanks of the three sets of conflicting SINE insertions (61/26/19) imply considerable support for the respective topologies of the SINEs. The 11,532 randomly selected sequence loci (each 2500 nt) revealed two conflicting tree topologies depending on the method applied (MP vs. ML). The IQ-TREE 2 gene concordance factor (gCF) and site concordance factor (sCF) for flanking and the 11,532 random sequences are shown below the bootstrap results. Bootstrap values are represented in bold.
). Sequence flanks of the three sets of conflicting SINE insertions (61/26/19) imply considerable support for the respective topologies of the SINEs. The 11,532 randomly selected sequence loci (each 2500 nt) revealed two conflicting tree topologies depending on the method applied (MP vs. ML). The IQ-TREE 2 gene concordance factor (gCF) and site concordance factor (sCF) for flanking and the 11,532 random sequences are shown below the bootstrap results. Bootstrap values are represented in bold.