Table 1.
Results obtained for viral taxonomic classification task regarding the genome type, realm, kingdom, phylum, class, order, family, and genus using XGBoost classifier. The features used were the genome’s sequence length (SL), the GC-content (GC), and the normalized compression (NC) values for the best model, the same model with IR configuration to 0, 1, and 2. The results correspond to the accuracy (ACC) and the probability of a random sequence being correctly classified (phit) using a random classifier (phit(CRandom)).
| Classification | N. Classes | N. Samples |
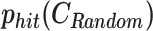
|
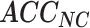
|
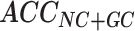
|
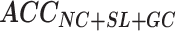
|
|
|
|---|---|---|---|---|---|---|---|---|
| Genome | 5 | 6089 | 20.00 | 75.57 | 80.60 | 87.11 | 81.24 | 87.25 |
| Realm | 5 | 5799 | 20.00 | 77.90 | 84.56 | 92.25 | 86.16 | 92.57 |
| Kingdom | 10 | 5788 | 10.00 | 76.44 | 82.51 | 90.82 | 84.06 | 90.96 |
| Phylum | 17 | 5778 | 5.88 | 63.97 | 70.69 | 82.36 | 73.21 | 83.41 |
| Class | 34 | 5845 | 2.94 | 59.83 | 65.90 | 79.05 | 68.66 | 80.23 |
| Order | 48 | 5838 | 2.08 | 58.44 | 65.08 | 78.20 | 67.88 | 79.62 |
| Family | 102 | 5990 | 0.98 | 43.35 | 54.06 | 72.46 | 58.34 | 74.46 |
| Genus | 360 | 4673 | 0.28 | 35.59 | 50.02 | 67.32 | 54.23 | 68.71 |
