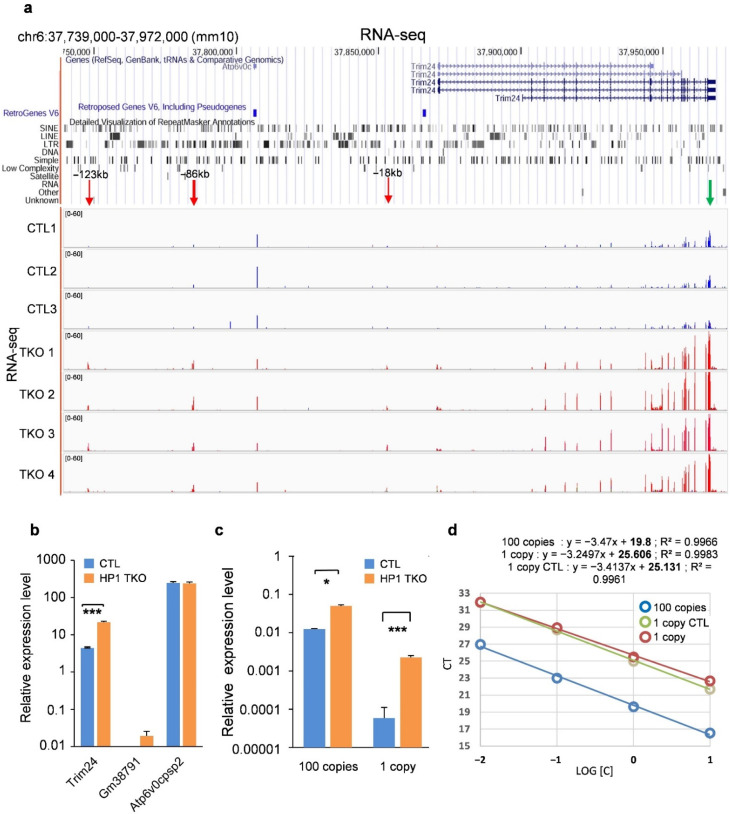
Figure 2.
HP1 proteins control the expression of Trim24 upstream ERV sequences. (a) Genome Browser snapshot of RNA-seq data at the Trim24 locus. Trim24 mRNA levels were found to be highly increased in the HP1-TKO compared to control livers. Upregulation was particularly clear for the last exons (green arrow on the right). One intergenic sequence, located 86 kb upstream of the Trim24 promoter, was found to be highly de-repressed in the HP1-TKO compared to control livers (large red arrow on the left). Two other regions, located 123 kb and 18 kb upstream of the Trim24 promoter, were also found to be de-repressed (faint red arrows on the left). (b) RT-qPCRs were performed on total RNA from three HP1-TKO (201, 248 and 316) and three control (202, 251 and 315) (CTL) mouse liver samples, using primers targeting Trim24 exon 19, Gm38791 and Atp6v0cpsp2 transcripts. RNA levels were normalised relative to Gapdh mRNA levels. We observe a significant difference in expression level between the two conditions only for Trim24. Error bars represent the s.e.m. of three biological replicates. p-value < 0.01 (***), (Student’s t-test). (c) Primer pairs binding to 1 copy of RLTR6-18 ERV or 100 copies of RLTR6-86 ERV Trim24 upstream sequences were used in RT-qPCR experiments and RNA levels were normalised relative to Gapdh mRNA levels. For both primer pairs, expression levels observed for the HP1-TKO was higher than in control mouse liver. Error bars represent s.e.m. of three biological replicates. p-value < 0.01 (***), and p-value < 0.05 (*) (Student t-tests). (d) Selection of primer pairs that amplify either one copy of the RLTR6-18 ERV (“1 copy”) or a hundred copies (“100 copies”) of ERV sequences in the mouse genome. The number of copies amplified by the selected primer pairs was determined on serial dilutions of genomic DNA (standard curves) in comparison with a control qPCR primer pair (“1 copy CTL”) that targets exon 1 of the Krüppel-like factor 4 (Klf4) gene, which amplifies exactly one copy in the mouse genome (see Materials and Methods).