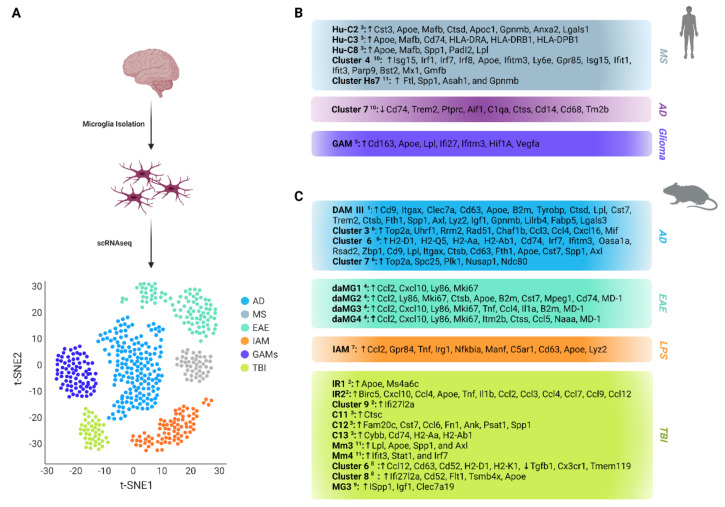
Figure 1.
Schematic overview of the most distinct disease-associated microglial populations and their transcriptomic signatures revealed through transcriptional profiling of murine and human microglia using single-cell RNA sequencing. (A) Schematic representation of research process for molecular classification of microglial subpopulations. (B) Most regulated human disease-associated microglial populations and their transcriptomic signatures. (C) Most regulated mouse disease-associated microglial populations and their transcriptomic signatures. Microglial subsets are represented with superscript numbers as described in the studies of 1 Keren-Shaul et al. [14], 2 Hammond et al. [15], 3 Masuda et al. [16], 4 Jordão et al. [17], 5 Sankowski et al. [18], 6 Mathys et al. [19], 7 Sousa et al. [20], 8 Wicher et al. [21], 9 Li et al. [22], 10 Olah et al. [23], and 11 Miedema et al. [24]. AD: Alzheimer’s Disease, MS: Multiple Sclerosis, EAE: Experimental Autoimmune Encephalomyelitis, LPS: Lipopolysaccharide, IAM: Inflammatory-Associated Microglia, GAM: Glioma-Associated Microglia HIV: Human Immunodeficiency Virus, TBI: Traumatic Brain Injury. ↑ denotes transcriptomic upregulation, ↓ denotes transcriptomic downregulation. Figure created with BioRender.com (accessed on 22 June 2022).