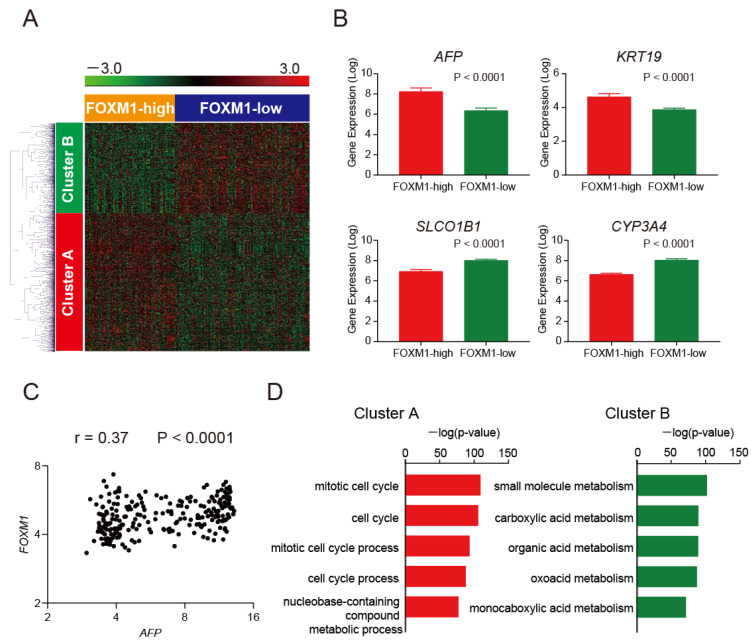
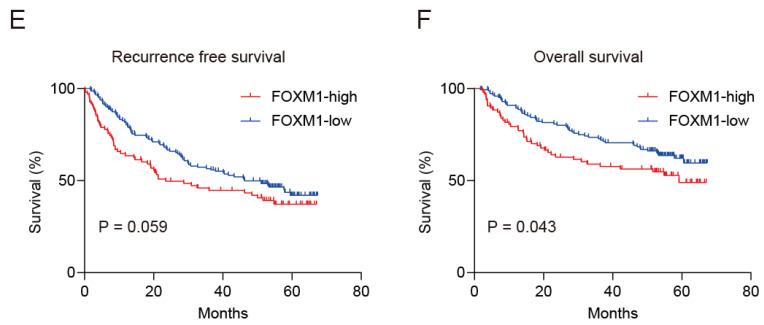
Figure 2.
Transcriptomic characteristics of FOXM1-high and -low HCC (Cohort 2). (A) Hierarchical cluster analysis of 2119 genes differentially expressed between FOXM1-high and -low HCC. A total of 1275 genes and 844 genes were upregulated or downregulated, respectively, in FOXM1-high HCC compared with -low HCC (p < 0.001). (B) Signal intensity of probes corresponding to AFP, KRT19, SLCO1B1, and CYP3A4 in FOXM1-high (red bar) and -low HCC (green bar) (Mann–Whitney test, means ± SEM). (C) Scatter plots analysis of AFP and FOXM1 expression in HCC (Spearman’s rank correlation coefficient). (D) Pathway analysis of FOXM1 co-regulated genes. Mitotic cell cycle processes were activated in FOXM1-high HCC (cluster A, red bars), whereas mature hepatocyte metabolism processes were inactivated in FOXM1-low HCC (cluster B, green bars). Significant processes are shown. (E) Kaplan–Meier survival analysis with the log-rank test of recurrence-free survival in FOXM1-high and -low HCC. Red, FOXM1-high (n = 94); blue, FOXM1-low (n = 143). (F) Kaplan–Meier survival analysis with the log-rank test of overall survival in FOXM1-high and -low HCC. Red, FOXM1-high (n = 94); blue, FOXM1-low (n = 143).