Abstract
Microsatellite instability (MSI) has been identified in several tumors arising from either germline or somatic aberration. The presence of MSI in cancer predicts the sensitivity to immune checkpoint inhibitors (ICIs), particularly PD1/PD-L1 inhibitors. To date, the predictive role of MSI is currently used in the selection of colorectal cancer patients for immunotherapy; moreover, the expansion of clinical trials into other cancer types may elucidate the predictive value of MSI for non-colorectal tumors. In clinical practice, several assays are used for MSI testing, including immunohistochemistry (IHC), polymerase chain reaction (PCR) and next-generation sequencing (NGS). In this review, we provide an overview of MSI in various cancer types, highlighting its potential predictive/prognostic role and the clinical trials performed. Finally, we focus on the comparison data between the different assays used to detect MSI in clinical practice.
Keywords: microsatellite instability, mismatch repair, immunotherapy, immunohistochemistry, PCR, NGS
1. Introduction
Microsatellites (MSs) are repeated sequences of 1–6 nucleotides in the human genome. MSs are highly polymorphic; in particular, their distribution and their dimensions are variable [1,2]. MSs are widely used both as markers in studies of genetic disease and as predictive biomarkers in various cancer types [3,4,5]. MSs are often subject to the incorporation or deletion of repeated units made by DNA polymerase during DNA replication; the mismatch repair system (MMR) is generally able to correct these errors [6]. The MMR is a pivotal system for maintaining genome integrity, consisting of several proteins, such as MLH1, MLH3, MSH2, MSH6, MSH3, PMS2, PMS1 and Exo1. The DNA errors are detected by MSH2/MSH6 and MSH2/MSH3 heterodimers; subsequently, the MLH1/PMS2 complex nicks DNA at sites of mismatch in order to begin strand repair [7]. MLH1 and MSH2 are the primary partners of their heterodimer, while PMS2 and MSH6 are the respective secondary partners. Consequently, the loss of the primary partners results in the loss of the entire heterodimer, but not the opposite. The alteration of the MMR proteins leads to a biological status known as microsatellite instability (MSI) [8]. The generation of MSI can be caused by the point mutations occurring in MMR genes, the DNA polymerase slippage during the replication process and the insertion/deletion of one or more bases in the microsatellite regions [1]. Furthermore, the MSI status could be due to other aberrations, including the hypermethylation of the MLH1 promoter [9], the epigenetic inactivation of MSH2 or MLH1 [10,11], the downregulation of MMR genes by microRNAs [12] or the slipped strand mating error (SSM) [13]. The MSI phenotype could be related to Lynch syndrome (LS), a genetic disease characterized by inherited germline mutations in MMR genes that is mainly shown in colorectal cancer and endometrial cancer; however, it is also shown in ovarian cancer, gastric cancer, the hepatobiliary tract, upper urinary tract, pancreas, brain and skin [14,15,16,17]. The presence of MSs in the coding regions is prone to generate genetic instability, leading to carcinogenesis [18]. Oda et al. proposed MSI classification into type A, with a variation of <6 bp, and type B, with a variation of >8 bp. Thibodeau et al. classified MSI based on the increase or decrease in fragment size into type I and II, respectively [19,20]. MSI was identified for the first time in colorectal cancer (CRC) [21]; subsequently, it was reported in endometrial cancer, ovarian cancer and gastric cancer (Figure 1). In recent years, MSI has also been identified with less frequency in other tumors, such as non-small cell lung cancer, breast cancer, brain cancer, prostate cancer, pancreatic cancer, bladder cancer and melanoma [22,23,24,25,26]. The immunocheckpoint inhibitors (ICI) have been shown to be effective in several tumor types carrying MSI-H compared to chemotherapy [27,28,29]. The rationale behind the sensitivity to ICI of MSI-H tumors is probably due to the accumulation of mutations that determines new mutated peptides, leading to the activation of the immune system [30]. In this context, MSI tumors have a high level of T-helper 1/cytotoxic lymphocytes and immune checkpoint molecules such as CTLA4, PD-1, PD-L1, LAG-3 and IDO, compared to MSS tumors [31,32]. The tumors carrying MSI showed a much better response to ICI than MSS cancers [33]. In recent years, the immunotherapy has achieved excellent results in several cancer types with the MSI phenotype [34]. In 2017, the US Food and Drug Administration (FDA) approved pembrolizumab, a PD-1 inhibitor, for the treatment of MSI advanced solid tumors regardless of the cancer type and the histology. MSI is the first cancer type-agnostic biomarker approved to establish the eligibility of patients for treatment with ICI rather than the site of the tumor [27].
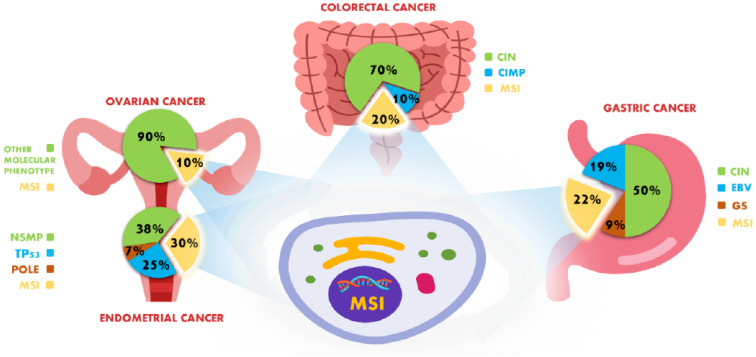
Figure 1.
Frequency of microsatellite instability in gastrointestinal and gynecologic cancers. Distribution of MSI according to the molecular classification of colorectal cancer, gastric cancer, ovarian and endometrial cancers. CIN: chromosome instability; CIMP: CpG island methylator phenotype; MSI: microsatellite instability; EBV: Epstein–Barr virus; GS: genomically stable; NSMP: no specific molecular profile; POLE: DNA polymerase epsilon.
2. Methods of MSI Detection
The MSI implication in cancer leads researchers to develop several techniques for its detection. The MSI phenotype could be defined both by a molecular approach evaluating the microsatellite sequences and by IHC evaluating the expression of MMR proteins (MMRP). The immunohistochemistry (IHC) analyzes the expression of a limited number of MMR proteins, including MLH1, PMS2, MSH2 and MSH6. MLH1 and MSH2 are the primary partners, and their degradation leads to the loss of their respective partners [35]. The proficient MMR (pMMR) cases show positive staining of the nuclei of all four MMRP; the deficient MMR (dMMR) cases show the loss of one of the two MLH1/PMS2 or MSH2/MSH6 heterodimers; the patchy MMR cases show focal IHC staining or cytoplasmic staining of one or more MMRP [36,37]. Some studies suggest that IHC could be exclusively used as a screening method for MSI detection since it is fast, easy to perform, inexpensive and widespread to most pathological laboratories [38,39,40,41]. Previous findings showed that both technical and interpretative pitfalls affect the MMR IHC, leading to false IHC results [42]. MMR IHC may be affected by pre-analytical problems, particularly the artifacts due to inadequate fixation. The adequate IHC staining interpretation requires the evaluation of a positive internal control, such as a normal stroma, and external control tissue, such as intratumoral lymphocytes and fibroblasts [43,44]. Moreover, a heterogeneous MMR IHC staining pattern could be caused by non-truncating mutations (missense mutations or frame insert/delete mutations) that could lead to full-length MMRP not functioning [41,45].
MSI molecular testing is generally performed by polymerase chain reaction (PCR). MSI-PCR is currently performed through two commercial panels, the Bethesda and the Pentaplex. The Bethesda panel includes five predefined genomic regions: two single-nucleotide (BAT25, BAT-26) and three dinucleotides (D2S123, D5S346 and D17S250). The Pentaplex panel includes five single-nucleotide loci, including BAT-25, BAT-26, NR-21, NR-24 and NR-27 [46,47]. The microsatellite status can be classified according to the number of instability markers, as follows: (i) high microsatellite instability (MSI-H) when there are more than 2 unstable loci; (ii) low microsatellite instability (MSI-L) when there is only one unstable locus; and (iii) microsatellite stability (MSS) when all the microsatellites are stable [48]. MSI-L and MSS tumors have similar molecular and clinical features; thus, patients carrying MSI-L are classified as MSS and they are not sensitive to ICI compared to patients carrying MSI-H [1]. MSI-PCR is a highly sensitive technique that is able to identify the instability due to the non-truncating mutations generally associated with a false positive IHC staining [41].
To date, both IHC and PCR are used in the clinical practice of MSI testing to define patients who can benefit from ICI treatment. However, several data in the literature showed discordant results between IHC and PCR in MSI detection. Several studies demonstrated that MMR IHC results frequently have not been confirmed by molecular analysis. For example, some cases of dMMR-IHC are results of MSS by PCR, resulting in clinical implications in the therapeutic choice. Likewise, cases with pMMMR-IHC were unexpectedly MSI by PCR [41,49,50,51,52]. These controversial results are generally attributable to the technical limits related to MMR IHC or a misinterpretation of the staining [41].
Recently, the next-generation sequencing (NGS) is another method used for MSI detection [2,53]. Several NGS-based methods have been described to assess MSI, including MSIseq somatic mutation analysis [54], mSINGS [53], MSISensor [55], Microsatellite Analysis for Normal Tumor Instability (MANTIS) [56], ColoSeq [57] and MSI-ColonCore [58]. The main advantage of an NGS-based approach is the simultaneous screening of microsatellite loci and other mutations including TMB [59].
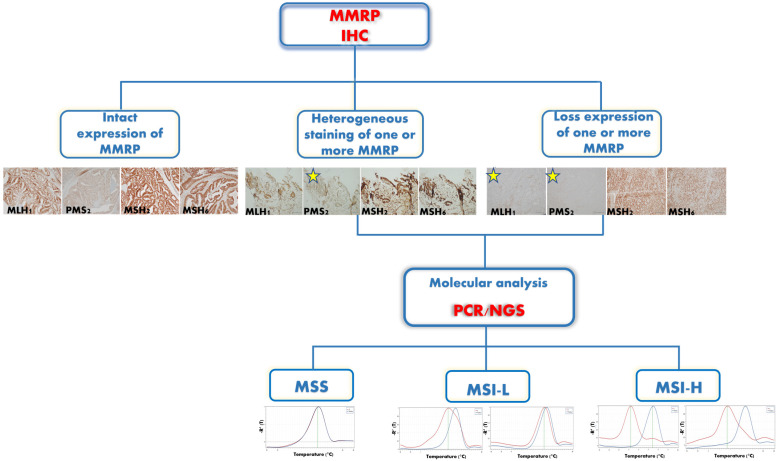
In conclusion, tumors with the loss or equivocal staining of at least one of the MMRP by IHC require an additional molecular test in order to avoid misdiagnosis and consequently inappropriate treatment choices [60] (Figure 2).
Figure 2.
The diagnostic algorithm of MSI status. MMRP IHC is used as a screening test: the intact expression of all four MMRPs is a conclusive result of the MSS status (original magnification 10×); the heterogeneous staining of one or more MMRPs needs an additional molecular assay (original magnification 10×); and the loss of one or more MMRPs needs a molecular assay (original magnification 10×). The molecular analysis performed through PCR and/or NGS leads to a conclusive result in cases with IHC equivocal staining. MMRP: mismatch repair proteins; IHC: immunohistochemistry; PCR: polymerase chain reaction; NGS: new-generation sequencing; MSS: microsatellite stability; MSI-L: microsatellite instability-low; MSI-H: microsatellite instability-high. The stars indicate the PMS2 heterogeneous staining (center) and the loss of MLH1 and PMS2 (right).
3. MSI in Colorectal Cancer
The Cancer Genome Atlas Network (TCGA) has included the MSI status in the molecular classification of CRC. Particularly, the TCGA classified CRC in different molecular subsets, including the chromosome instability phenotypes (CIN), accounting for 70% of cases, the MSI phenotype, accounting for 20%, and the CpG island methylation phenotype (CIMP), accounting for 10% [61] (Figure 1). Although some studies showed that some MSI CRC cases harbouring also CIMP and CIN overlap with different molecular phenotypes, TCGA classified MSI CRC as a distinct molecular phenotype [62,63]. Approximately 3% to 5% of all CRCs have dMMR associated with LS. This syndrome, known as hereditary non-polyposis colorectal carcinoma (HNPCC), is an autosomal dominant syndrome with germline mutations of the MMR genes [64,65,66]. Beyond the CRCs associated with LS, approximately 70–95% of MSI CRCs are sporadic [67]. The relationship between CRC MSI and some clinical features, such as primary right-sided tumors and older age, has been demonstrated [68]. The MSI status is an early event in the development of CRC; it has never been identified in polyps [69]. On the contrary, MSI has also been observed in the normal colonic mucosa of patients with LS [70,71,72]. Several studies suggest that MSI CRCs at advanced stages show a poor prognosis compared to MSS CRC [73,74]. The clinical studies KEYNOTE-016 and KEYNOTE-164 demonstrated the efficacy of pembrolizumab, with durable responses in patients with advanced MSI-H/dMMR or mCRC treated after chemotherapy [75,76]. In KEYNOTE-177, a phase 3 study, patients with unresectable or metastatic MSI/dMMR CRC were treated with pembrolizumab as the first-line treatment without receiving prior therapy; this showed clinically significant improvements in survival compared to chemotherapy. In 2020, the FDA approved pembrolizumab as the first-line treatment for advanced or metastatic CRC with MSI-H/dMMR [77]. Furthermore, CheckMate 142 demonstrated the efficacy of nivolumab (an anti-PD1 antibody) used alone or in combination with ipilimumab (an anti-CTLA-4 antibody) in mCRC MSI-H/dMMR patients previously treated with chemotherapy. In 2018, the FDA approved nivolumab either alone or in combination with ipilimumab [78,79] (Table 1). The predictive value of MSI in CRC requires the optimization of testing in clinical practice. As already mentioned, IHC is the most used method; however, it showed several discrepancies with PCR and NGS results. Several studies have compared the different assays for MSI detection in CRC series [41,50,80,81,82] (Table 2). The comparative analysis between different methods showed several discrepancies; in particular, some of the dMMR-IHC cases were MSS-PCR (ranging from 1% to 33% of cases) [82,83,84]. In contrast, only 5% of pMMR-IHC cases analyzed in the literature showed MSI by PCR [41,50,80,81,85]. Generally, the PCR results are in agreement with the NGS results, showing a discrepancy of up to 3% [58,83,86,87]. To date, no recommendations have been proposed regarding the gold standard molecular test for MSI analysis. The NGS is the most sensitive method for MSI detection. Moreover, the NGS represents the best molecular approach since it allows simultaneously the detection of both MSI and a wide panel of other mutations. However, PCR can represent a valid molecular test for MSI detection in daily clinical practice since it is economical and widespread in the laboratories compared to NGS. Finally, MSI analysis by NGS could be useful in selected cases showing discordant results between IHC and PCR.
Table 1.
Clinical trials for immune checkpoint inhibitors treatment of MSI CRC.
| Clinical Trial | Drug Treatment | Phase | Tumor Type | N. Patients |
Status | Conclusion |
|---|---|---|---|---|---|---|
|
NCT02460198 KEYNOTE-164 |
Pembrolizumab | 2 | Advanced unresectable CRC mCRC Stage IV |
124 | Completed | Pembrolizumab is effective in MSI-H/dMMR CRC |
|
NCT01876511 KEYNOTE-016 |
Pembrolizumab | 2 | MSI-H CRC MSS CRC MSI non-CRC |
113 | Completed | ORR: disappearance of all target lesions PR: 30% decrease in the diameters of the target lesions |
| NCT00912743 | Olaparib | 2 | CRC MSI-H | 33 | Completed | Olaparib activity not demonstrated after failure of standard systemic therapy |
|
NCT03374254 KEYNOTE-651 |
Pembrolizumab + Binimetinib alone Pembrolizumab + Chemotherapy with or without Binimetinib |
1 | mCRC MSS/pMMR | 220 | Active, not recruiting | No results posted |
|
NCT02060188 CheckMate 142 |
Nivolumab alone vs. Nivolumab + Ipilimumab orNivolumab + BMS-986016 or Nivolumab + Daratumumab Nivolumab + Ipilimumab + Cobimetinib |
2 | mCRC MSI-H CRC non-MSI-H |
385 | Active, not recruiting | No results posted |
| NCT04258111 | IBI310 (anti-CTLA-4 antibody) + Sintilimab (anti-PD-1 antibody) | 2 | CRC MSI-H mCRC MSI-H |
4 | Active, not recruiting | No results posted |
| NCT03435107 | Durvalumab | 2 | mCRC MSI-H or POLE | 33 | Active, not recruiting | No results posted |
| NCT03350126 | Ipilimumab + Nivolumab | 2 | mCRC MSI/dMMR | 57 | Active, not recruiting | No results posted |
| NCT03186326 | FOLFOX or FOLFIRI Protocol + Panitumumab + Cetuximab + Bevacizumab + Aflibercept vs. Avelumab |
2 | mCRC MSI-H | 132 | Active, not recruiting | No results posted |
|
NCT02563002 KEYNOTE-177 |
Pembrolizumab vs. standard therapy (mFOLFOX6 and FOLFIRI alone or associated with Bevacizumab + Cetuximab) | 3 | MSI CRC stage IV | 307 | Active, not recruiting | Pembrolizumab prolongs PFS or OS |
| NCT03827044 | Avelumab | 3 | POLE CRC CRC MSI Stage III |
402 | Active, not recruiting | No results posted |
| NCT03926338 | Toripalimab + Celecoxib vs. Toripalimab alone |
1–2 | MSI-H/dMMR CRC | 34 | Recruiting | No results posted |
| NCT04636008 | Sintilimab + Hypofractionated radiotherapy |
1–2 | Rectal cancer MSI-H/dMMR | 20 | Recruiting | No results posted |
| NCT04014530 | Ataluren + Pembrolizumab | 1–2 | mCRC pMMR and dMMR mEC dMMR |
47 | Recruiting | No results posted |
| NCT04988191 | Toripalimab + Bevacizumab + Irinotecan | 1–2 | CRC dMMR/MSI-H | 44 | Recruiting | No results posted |
| NCT04715633 | Camrelizumab + Apatinib | 2 | Locally advanced dMMR/MSI-H CRC | 52 | Recruiting | No results posted |
| NCT03519412 | Temozolomide (induction) Pembrolizumab (treatment) |
2 | mCRC pMMR and dMMR | 102 | Recruiting | No results posted |
| NCT05116085 | Tislelizumab | 2 | With early-stage (Stage II-III) MSI-H or dMMR CRC | 38 | Recruiting | No results posted |
| NCT04715633 | Camrelizumab + Apatinib | 2 | MSI-H/dMMR CRC | 52 | Recruiting | No results posted |
| NCT05118724 | Atezolizumab | 2 | CRC MSI-H/dMMR Stage III | 120 | Recruiting | No results posted |
| NCT04695470 | Fruquintinib + Sintilimab | 2 | mCRC TMB and non MSI-H | 70 | Recruiting | No results posted |
| NCT03638297 | BAT1306 + Aspirin | 2 | CRC MSI-H/dMMR or TMB | 27 | Recruiting | No results posted |
| NCT04730544 | Nivolumab + Ipilimumab | 2 | mCRC dMMR/MSI-H | 96 | Recruiting | No results posted |
| NCT04895722 | Pembrolizumab alone vs. Pembrolizumab + Quavonlimab vs. Pembrolizumab + Favezelimab vs. Pembrolizumab + Vibostolimab |
2 | CRC MSI Stage IV | 320 | Recruiting | No results posted |
| NCT04301557 | PD-1 Antibody + Oxaliplatin + Capecitabine + External beam radiotherapy + Total mesorectal excision | 2 | Advanced CRC dMMR/MSI-H | 25 | Recruiting | No results posted |
| NCT04304209 | Oxaliplatin + Capecitabine + Sintilimab + Total mesorectal excision vs. Oxaliplatin + Capecitabine + Sintilimab + Radiotherapy + Total mesorectal excision vs. Oxaliplatin + Capecitabine + Radiotherapy + Total mesorectal excision |
2–3 | CRC Stage IICRC Stage III dMMR/MSI-H | 195 | Recruiting | No results posted |
| NCT02912559 | Atezolizumab + Fluorouracil vs. Leucovorin calcium + Oxaliplatin vs. Fluorouracil + Leucovorin calcium + Oxaliplatin |
3 | CRC dMMR Stage III | 700 | Recruiting | No results posted |
| NCT05239741 | Pembrolizumab alone vs. 5-fluorouracil alone or 5-fluorouracil + Bevacizumab or 5-fluorouracil + Cetuximab or FOLFIRI + Bevacizumab or FOLFIRI + Cetuximab |
3 | CRC MSI-H Stage IV dMMR CRC |
100 | Recruiting | No results posted |
| NCT02997228 | Atezolizumab alone vs. mFOLFOX6 + Bevacizumab vs. mFOLFOX6 + Bevacizumab + Atezolizumab |
3 | mCRC MSI-H | 231 | Recruiting | No results posted |
| NCT04008030 | Nivolumab alone vs. Nivolumab + Ipilimumab vs. Active comparator (Oxaliplatin, Leucovorin, Fluorouracil, Irinotecan, Bevacizumab, Cetuximab) |
3 | mCRC MSI-H | 748 | Recruiting | No results posted |
| NCT05236972 | Sintilimab alone vs. Oxaliplatin + Capecitabine |
3 | CRC dMMR/MSI-H Stage III | 323 | Recruiting | No results posted |
| NCT05239741 | Pembrolizumab alone vs. Active comparator (Oxaliplatin, Leucovorin, 5-fluorouracil, Irinotecan, Bevacizumab, Cetuximab) |
3 | CRC MSI-H/dMMR Stage IV | 100 | Recruiting | No results posted |
| NCT05217446 | Pembrolizumab alone vs. Encorafenib + Cetuximab + Pembrolizumab |
2 | mCRC MSI-H/dMMR | 104 | Not yet recruiting |
No results posted |
| NCT05217446 | Pembrolizumab alone vs. Encorafenib + Cetuximab + Pembrolizumab |
2 | mCRC MSI-H/dMMR | 104 | Not yet recruiting |
No results posted |
| NCT05231850 | Tislelizumab | 2 | CRC dMMR/MSI-H Stage II and III | 70 | No yet recruiting |
No results posted |
| NCT05231850 | Tislelizumab | 2 | CRC dMMR/MSI-H Stage II and Stage III | 70 | Not yet recruiting |
No results posted |
| NCT04866862 | Fruquintinib + Camrelizumab | 2 | Non MSI-H/dMMR Refractory CRC | 32 | Not yet recruiting |
No results posted |
| NCT05131919 | Pembrolizumab | 2 | Locally advanced, Irresectable, dMMR not- mCRC | 25 | Not yet recruiting |
No results posted |
| NCT05215379 | Xintilimab injection | 2–3 | Rectal cancer immunotherapy MSI-L |
180 | Not yet recruiting |
No results posted |
CRC: colorectal cancer; mCRC: metastatic CRC; MSS: microsatellite stability; MSI: microsatellite instability; MSI-H: microsatellite instability high; MSI-L: microsatellite instability low; dMMR: deficient mismatch repair; TMB: tumor mutational burden; POLE: DNA polymerase epsilon; ORR: objective response rate; PFS: progression free survival; PR: partial response; CR: complete response; RECIST: response evaluation criteria in solid tumors.
Table 2.
Comparison between IHC, PCR and NGS in CRC cases.
| Cases | IHC Results | PCR Results | NGS Results | References |
|---|---|---|---|---|
| 28 | 16 dMMR | 15 MSI-H | 15 MSI | [83] |
| 1 MSS | 1 MSS | |||
| 12 pMMR | 12 MSS | 12 MSS | ||
| 93 | 135 dMMR | 132 MSI-H | NP | [84] |
| 3 MSS | ||||
| 458 pMMR | 4 MSI-H | |||
| 10 MSI-L | ||||
| 444 MSS | ||||
| 988 | 102 dMMR | 98 MSI-H | NP | [41] |
| 4 MSS/MSI-L | ||||
| 886 pMMR | 4 MSI-H | |||
| 882 MSS/MSI-L | ||||
| 91 | 54 dMMR | 48 MSI-H | 47 MSI | [58] |
| 1 MSS | ||||
| 6 MSS/MSI-L | 6 MSS | |||
| 37 pMMR | 37 MSS/MSI-L | 37 MSS | ||
| 73 | 12 dMMR | 8 MSI-H | NP | [50] |
| 1 MSI-L | ||||
| 3 MSS | ||||
| 61 pMMR | 3 MSI-H | |||
| 11 MSI-L | ||||
| 47 MSS | ||||
| 15 | 6 dMMR | 6 MSI-H | 6 MSI | [86] |
| 1 loMMR | 1 MSI-H | 1 MSI | ||
| 1 paMMR | 1 MSS | 1 MSS | ||
| 6 pMMR | 6 MSS | 1 MSI | ||
| 5 MSS | ||||
| 1 NV | 1 MSS | 1 MSS | ||
| 262 | 28 dMMR | 26 MSI-H | NP | [80] |
| 2 MSI-L | ||||
| 234 pMMR | 9 MSI-L | |||
| 225 MSS | ||||
| 296 | 65 dMMR | 63 MSI-H | NP | [82] |
| 2 MSS | ||||
| 7 loMMR | 5 MSI-H | |||
| 2 MSI-L | ||||
| 224 pMMR | 7 MSI-L | |||
| 217 MSS | ||||
| 98 | 38 dMMR | 33 MSI-H | 32 MSI-H | [85] |
| 1 MSS | ||||
| 1 MSI-L | 1 MSS | |||
| 4 MSS | 4 MSS | |||
| 60 pMMR | 1 MSI-H | 1 MSS | ||
| 59 MSS | 59 MSS | |||
| 809 | 148 dMMR | 147 MSI-H | NP | [81] |
| 1 MSS | ||||
| 12 loMMR | 11 MSI-H | |||
| 1 MSS | ||||
| 649 pMMR | 1 MSI-H | |||
| 3 MSI-L | ||||
| 645 MSS | ||||
| 166 | 75 dMMR | 75 MSI-H | NP | [87] |
| 91 pMMR | 90 MSS/MSI-L | |||
| 1 MSI-H |
dMMR: deficient mismatch repair; pMMR: proficient mismatch repair; IHC: immunohistochemistry; PCR: polymerase chain reaction; NGS: next generation sequencing; MSI-H: microsatellite instability-high; MSS: microsatellite stability; MSI-L: microsatellite instability-low; NP: not performed.
4. MSI in Gastric Cancer
The TCGA has included MSI status in the molecular classification of GC. Particularly, the TCGA classified GC in different molecular subsets, including the chromosome instability phenotypes (CIN), accounting for 50% of cases, the genomically stable (GS) tumors, accounting for 19%, the MSI phenotype, accounting for 22%, and the Epstein-Barr virus (EBV)-positive tumors, accounting for 9% (Figure 1) [88]. Although rare cases simultaneously carrying MSI and EBV have been identified, the TCGA considers MSI as a molecular profile distinct from the others [89]. Similarly, the Asian Cancer Research Group (ACRG) classified GC in MSS and MSI [90]. Many studies have found MSI in precancerous lesions and an increase in MSI frequency in GC cases is observed [91,92]. MSI GCs usually occur in older female patients, with an intestinal subtype at an early stage (I/II) and with better differentiation and distal position, and no lymph node involvement [44,91]. Ottini et al. observed that the MLH1 promoter methylation is frequently in sporadic GC, while MLH1 and MSH2 mutations are rare in MSI GCs [93]. MSI occurs both in sporadic GCs and in GCs associated with LS [94,95]. A positive correlation was observed between the presence of Helicobacter pylori (H. pylori) and MSI. The active bacterial infection is present more frequently in GC MSI patients, suggesting that H. pylori may affect the MMR system during the gradual progression of gastric carcinogenesis [52]. Approximately 85% of the MSI GC cases concomitantly showed a KRAS mutation. On the contrary, no association between MSI GC and a mutated BRAF was reported [96]. MSI and human epidermal growth factor receptor 2 (HER2) amplification are reported to be mutually exclusive, suggesting that HER2 amplification could be a negative predictive marker for immunotherapy in GC patients [97]. The MSI status is not used as an exclusive prognostic biomarker, since the prognosis of GC patients is affected by other clinical and pathological features, including the age, stage, grade and chemotherapy treatment [98]. To date, controversial data about the prognostic role of MSI in GC have been reported. Several studies have shown that GCs carrying MSI have a good prognosis [91,99,100]. However, other studies showed MSI not having a prognostic role; in particular, MSI GCs do not have a better survival time than MSI-L/MSS patients. [101,102]. The metastatic MSI GCs have shown long-lasting positive results after receiving treatment with anti-PD-1 antibodies (durvalumab, pembrolizumab and nivolumab), as a single therapy or in combination with anti-CTLA4 antibodies (ipilimumab and tremelimumab) [103]. Pembrolizumab was investigated among patients with advanced MSI gastric or gastroesophageal junction cancer who were enrolled in different studies, such as KEYNOTE-059, KEYNOTE-061 and KEYNOTE-062. The results indicate that pembrolizumab or pembrolizumab plus chemotherapy provided durable antitumor activity compared to chemotherapy alone in patients with an MSI gastric junction or gastroesophageal cancer. Other clinical trials have suggested drug combinations, such as durvalumab and tremelimumab or nivolumab and ipilimumab in phase 2, for the treatment of advanced MSI GC [104,105,106] (Table 3). The comparative analysis between these methods showed some discrepancies, particularly as some dMMR-IHC cases were MSS by PCR (ranging from 1% to 38%) [51,107], while pMMR-IHC cases were MSI by molecular analysis (ranging from 1% to 7,4%) [52,108,109].
Table 3.
Clinical trials for immune checkpoint inhibitors treatment of MSI GC.
| Clinical Trials | Drug Treatment |
Phase | Tumor Type | N. Patients |
Status | Conclusions |
|---|---|---|---|---|---|---|
| NCT04795661 | Pembrolizumab | 2 | Localized resectable tumor MSI/dMMR or EBV-positive GC | 120 | Recruiting | No results posted |
| NCT04817826 | Durvalumab + Tremelimumab | 2 | GC MSI-H | 31 | Recruiting | No results posted |
| NCT05177133 | Capecitabine + Oxaliplatin + Retifanlimab | 2 | dMMR Esophagogastric cancer | 25 | Recruiting | No results posted |
| NCT04152889 | Camrelizumab + S-1 + Docetaxel | 2 | GC Stage III (PD-L1 +/MSI-H/EBV +/dMMR) | 20 | Recruiting | No results posted |
| NCT04006262 | Nivolumab + Ipilimumab | 2 | Localized MSI and/or dMMR Oeso-gastric adenocarcinoma | 32 | Recruiting | No results posted |
| NCT03257163 | Capecitabine + Pembrolizumab + Radiation therapy | 2 | dMMR and Epstein–Barr virus Positive GC | 40 | Recruiting | No results posted |
GC: gastric cancer; MSI: microsatellite instability; MSI-H: microsatellite instability-high; dMMR: deficient mismatch repair; EBV: Epstein–Barr virus; PD-L1: programmed death-ligand 1.
The FDA accelerated approval of pembrolizumab in 2017 to treat MSI patients in solid tumors, including GC, who have progressed following prior treatment [27]. Few data have been reported regarding the MSI testing in GC; all the comparison data between IHC and PCR are summarized in Table 4.
Table 4.
Comparison between IHC, PCR and NGS in GC cases.
| Cases | IHC Results | PCR Results | NGS Results | References |
|---|---|---|---|---|
| 56 | 13 dMRR | 8 MSI-H | NP | [107] |
| 5 MSI-L | ||||
| 43 pMMR | 43 MSS | |||
| 580 | 61 dMRR | 60 MSI-H | NP | [51] |
| 1 MSS | ||||
| 519 pMMR | 519 MSS | |||
| 60 | 6 dMRR | 6 MSI-H | NP | [52] |
| 54 pMMR | 4 MSI-H | |||
| 50 MSS | ||||
| 16 | 9 dMMR | 9 MSI-H | NP | [108] |
| paMMR | 1 MSI-H | |||
| lo-paMMR | 1 MSI-H | |||
| 5 pMMR | 5 MSS | |||
| 50 | 4 dMMR | 4 MSI-H | NP | [109] |
| 46 pMMR | 44 MSS | |||
| 2 MSI-H |
MMR: deficient mismatch repair; pMMR: proficient mismatch repair; IHC: immunohistochemistry; PCR: polymerase chain reaction; NGS: next generation sequencing; MSI-H: microsatellite instability-high; MSS: microsatellite stability; MSI-L: microsatellite instability-low; NP: not performed.
5. MSI in Gynecologic Cancers
Several studies have shown the presence of MSI from 2 to 20% in ovarian cancer (OC) (Figure 1), particularly in clear cell carcinomas and endometrioid cancers [110,111,112]. In clear cell carcinoma, the expression of CD8, PD-1 and tumor-infiltrating lymphocytes (TIL) was higher in MSI tumors than in those of MSS. In contrast, in endometrioid carcinomas, no correlation between the MSI status and TIL was reported [111,113]. MSI OCs are characterized by advanced stage, poor differentiation, grade and poor prognosis [114]. MSI OCs are frequently associated with LS [115]. However, the correlation between MSI and LS is not always verified since the LS was observed in both MSI OC and MSS OC [116]. MSI detection in OC has not yet been extensively studied and no adequate method is currently defined. The suitability and specificity of the Bethesda panel are still today an open issue in this cancer type [117]. There are few studies that have compared IHC and PCR for detecting MSI in OC (Table 5).
Table 5.
Comparison between IHC, PCR and NGS in OC cases.
| Cases | IHC Results | PCR Results | NGS Results | References |
|---|---|---|---|---|
| 30 | 3 dMMR | 3 MSI-H | NP | [113] |
| 27 pMMR | 27 MSS | |||
| 834 | 228 dMMR | 41 MSI-H | NP | [118] |
| 187 MSS | ||||
| 606 pMMR | 83 MSI-H | |||
| 523 MSS | ||||
| 26 | 7 dMMR | 7 MSI-H | NP | [119] |
| 19 pMMR | 1 MSI-H | |||
| 18 MSS | ||||
| 42 | 4 dMMR | 4 MSI-H | NP | [120] |
| 38 pMMR | 2 MSI-H | |||
| 3 MSI-L | ||||
| 33 MSS |
dMMR: deficient mismatch repair; pMMR: proficient mismatch repair; IHC: immunohistochemistry; PCR: polymerase chain reaction; NGS: next generation sequencing; MSI-H: microsatellite instability-high; MSS: microsatellite stability; MSI-L: microsatellite instability-low; NP: not performed.
A concordance was shown between the IHC analysis and PCR [113]. Some discordant results have been observed; in particular, dMMR-IHC cases were MSS by PCR in a wide subset accounting from 1% to 80% [118,119,120]. Although MSI is still a poorly understood marker in OC patients, several studies showed its potential predictive role for ICI in this cancer (Table 6). The TCGA has included the MSI status in the molecular classification of endometrial cancer (EC). Particularly, the TCGA classified EC in different molecular subsets, including the no specific molecular profile (NSMP), accounting for 38% of cases, the MSI phenotype, accounting for 30%, the copy number high/mutant TP53 (CNH), accounting for 25%, and the POLE/ultramutated (POLE), accounting for 7% (Figure 1) [121,122,123]. Although some studies showed that some MSI EC cases also harbouring other mutations overlapped with different molecular phenotypes, the TCGA classified MSI EC as a distinct molecular phenotype [124]. LS in the EC was observed with a frequency of 3% [125].
Table 6.
Clinical trials for immune checkpoint inhibitors treatment of MSI OC.
| Clinical Trial | Drug Treatment | Phase | Tumor Type | N. Patients | Status | Conclusion |
|---|---|---|---|---|---|---|
| NCT03836352 | DPX-Survivac + Cyclophosphamide + Pembrolizumab vs. DPX-Survivac + Pembrolizumab |
2 | Solid tumors, including OC and MSI-H | 184 | Active, not recruiting | No results posted |
OC: ovarian cancer; MSI-H: microsatellite instability-high.
Regarding morphological features, MSI ECs are frequently characterized by the presence of peritumoral lymphocytes and TILs [126]. In approximately 95% of ECs, the MSI is due to the hypermethylation of the MLH1 promoter [127]. MSI ECs showed in about 10–53% of cases the simultaneous presence of other gene alterations, particularly the mutations of the following genes: RPL22 (46%) [128,129,130], PTEN (34%) [128,131], KRAS (35%) [128], ATR (15%) [132], CHK1 (29%) [133], Caspase5 (10%) [131,134] and BAX (27%) [129,134,135]. Furthermore, the PTEN gene mutations, frequently observed in sporadic EC (25–83%), showed a correlation with MSI with a frequency of 60% [136]. Moreover, MSI ECs are associated with a higher neoantigen load and increased TIL, CD3-positive, CD8-positive and PD-L-1 positive compared to MSS/pMMR EC [137]. The role of MSI in ECs is until now unclear; most of the studies have concluded that the MSI status has no prognostic role [138,139,140]. Previous studies have shown that the combination of pembrolizumab and lenvatinib can be used for second-line treatment in patients with EC who are not dMMR/MSI-H, and studies are ongoing for the treatment of patients with advanced EC MSI [141,142]. In the USA, both pembrolizumab and dostarlimab are approved as monotherapy for the treatment of MSI EC patients. In the EU, dostarlimab is the only therapy approved for patients with EC dMMR/MSI who have failed platinum therapy [143] (Table 7). Several studies have compared the different assays for MSI detection in EC series (Table 8).
Table 7.
Clinical trials for immune checkpoint inhibitors treatment of MSI EC.
| Clinical Trial | Drug Treatment | Phase | Tumor Type | N. Patients |
Status | Conclusion |
|---|---|---|---|---|---|---|
| NCT04906382 | Carboplatin + Paclitaxel + Tislelizumab | 1 | dMMR EC | 20 | Recruiting | No results posted |
| NCT05112601 | Ipilimumab + Nivolumab vs. Nivolumab alone | 2 | dMMR recurrent EC | 12 | Recruiting | No results posted |
| NCT02912572 | Avelumab alone vs. Avelumab + Talazoparib vs. Avelumab + Axitinib |
2 | mEC MSI-H | 105 | Recruiting | No results posted |
| NCT04774419 | Intensity modulated radiation therapy (IMRT) + TSR-042 | 2 | EC dMMR/MSI-H | 31 | Recruiting | No results posted |
| NCT05036681 | Futibatinib vs. Pembrolizumab | 2 | MSS mEC | 30 | Recruiting | No results posted |
|
NCT05173987 Keynote C93 |
Pembrolizumab alone vs. carboplatin + paclitaxel + docetaxel + cisplatin |
3 | dMMR EC | 350 | Recruiting | No results posted |
| NCT03241745 | Nivolumab | 2 | EC dMMR/MSI-H | 35 | Active, not recruiting | No results posted |
| NCT05201547 | Dostarlimab alone vs. Carboplatin-Paclitaxel |
3 | EC dMMR | 142 | Not yet recruiting | No results posted |
EC: endometrial cancer; mCRC: metastatic EC; MSS: microsatellite stability; dMMR: deficient mismatch repair.
Table 8.
Comparison between IHC, PCR and NGS in EC cases.
| Cases | IHC Results | PCR Results | NGS Results | References |
|---|---|---|---|---|
| 108 | 33 dMMR | 27 MSI-H | NP | [144] |
| 6 MSS | ||||
| 75 pMMR | 75 MSS | |||
| 98 | 18 dMMR | 8 MSI-H | NP | [145] |
| 10 MSS/MSI-L | ||||
| 5 loMMR | 2 MSI-H | |||
| 3 MSS/MSI-L | ||||
| 75 pMMR | 75 MSS | |||
| 100 | 52 dMMR | 51 MSI-H | NP | [146] |
| 1 MSS | ||||
| 10 loMMR | 6 MSI-H | |||
| 4 MSS | ||||
| 9 paMMR | 3 MSI-H | |||
| 1 MSI-L | ||||
| 5 MSS | ||||
| 18 pMMR | 18 MSS | |||
| 11 NA | 8 MSI-H | |||
| 3 MSS | ||||
| 89 | 26 dMMR | 23 MSI-H | NP | [147] |
| 3 MSS | ||||
| 63 pMMR | 3 MSI-H | |||
| 60 MSS | ||||
| 99 | 29 dMMR | NA | 16 MSI | [148] |
| 13 MSS | ||||
| 70 pMMR | 2 MSI | |||
| 68 MSS | ||||
| 21 | 9 dMMR | 6 MSI-H | 8 MSI | [83] |
| 2 MSS | ||||
| 1 MSI-L | 1 MSS | |||
| 3 loMMR | 2 MSS | 2 MSS | ||
| 1 MSI-H | 1 MSI | |||
| 9 pMMR | 9 MSS | 9 MSS | ||
| 15 | 6 dMMR | 5 MSI-H | 6 MSI | [86] |
| 1 NA | ||||
| 3 loMMR | 2 MSI-H | 1 MSI | ||
| 1 MSS | ||||
| 1 MSS | 1 MSS | |||
| 1 paMMR | 1 MSS | 1 MSS | ||
| 5 pMMR | 5 MSS | 5 MSS |
dMMR: deficient mismatch repair; pMMR: proficient mismatch repair; IHC: immunohistochemistry; PCR: polymerase chain reaction; NGS: next generation sequencing; MSI-H: microsatellite instability-high; MSS: microsatellite stability; MSI-L: microsatellite instability-low; NA: not available; NP: not performed.
The comparative analysis between these methods showed some discrepancies; in particular, approximately 1–68% of dMMR-IHC cases were MSS by PCR/NGS [83,86,144,145,146], and about 5% of pMMR-IHC cases were MSI-H by molecular analysis [147,148].
6. MSI in Other Malignancies
MSI has been found in several cancer types, including non-small cell lung cancer, melanoma, breast cancer, urothelial cancer, pancreatic ductal adenocarcinoma and brain cancer. The expansion of MSI clinical trials into other cancers may elucidate the prognostic and predictive value of MSI for non-colorectal. The frequency of MSI reported in non-small cell lung cancer (NSCLC) is very heterogeneous, ranging from 0.8% to 40% [149,150,151,152]. The role and clinical implications of MSI are still unclear in NSCLC. Carpagnano et al. suggested that MSI is associated with a poor prognosis in NSCLC [153].
Moreover, recent findings suggested that MSI in NSCLC is associated with an increased response to immunotherapy [154]. Several clinical trials showed the efficacy of pembrolizumab for the treatment of previously treated NSCLC patients carrying MSI [155,156,157] (Table 9).
Table 9.
Clinical trials for immune checkpoint inhibitors treatment of other MSI/no-MSI-H tumors.
| Clinical Trial | Drug Treatment | Phase | Tumor Type | N. Patients |
Status | Conclusion |
|---|---|---|---|---|---|---|
|
NCT01876511 KEYNOTE-016 |
Pembrolizumab | 2 | MSI CRC MSS CRC MSI Non-CRC |
113 | Completed | ORR: disappearance of all target lesions; PR: 30% decrease in the diameters of the target lesions. |
| NCT04328740 | TP-1454 monotherapy vs. TP-1454 + Ipilimumab + Nivolumab |
1 | Advanced solid tumor Advanced/metastatic RCC, MSI-H or dMMR mCRC, NSCLC | 44 | Recruiting | No results posted |
|
NCT02332668 KEYNOTE-051 |
Pembrolizumab | 1–2 | Melanoma, Lymphoma, Solid tumor, Classical Hodgkin Lymphoma; MSI-H solid tumor |
320 | Recruiting | No results posted |
| NCT04521075 | Fecal microbial transplantation by capsules | 1–2 | Metastatic or inoperable melanoma, MSI-H, dMMR or NSCLC | 42 | Recruiting | No results posted |
| NCT03607890 | Nivolumab + Relatlimab | 2 | Advanced dMMR cancers resistant to Prior PD-(L)1 inhibitor | 42 | Recruiting | No results posted |
| NCT03667170 | Envafolimab | 2 | dMMR/MSI-H advanced solid tumors | 200 | Recruiting | No results posted |
| NCT03236935 | L-NMMA + Pembrolizumab | 1 | NSCLC, Malignant melanoma Head and neck squamous cell carcinoma Classical Hodgkin Lymphoma Urothelial carcinoma Bladder DNA repair-deficiency disorders |
12 | Active, not recruiting | No results posted |
| NCT03053466 | APL-501 | 1 | Solid tumors MSI-H or dMMR | 30 | Active, not recruiting | No results posted |
| NCT04800627 | Pembrolizumab + Pevonedistat | 1–2 | dMMR/MSI-H metastatic or locally advanced unresectable solid tumor | 2 | Active, not recruiting | No results posted |
| NCT02983578 | Danvatirsen + Durvalumab | 2 | Advanced and refractory pancreatic, NSCLC and dMMR CRC | 53 | Active, not recruiting | Not result posted |
| NCT03241745 | Nivolumab | 2 | MSI/dMMR/Hypermutated uterine cancer | 35 | Active, not recruiting | No results posted |
| NCT04326829 | QL1604 | 2 | dMMR or MSI-H advanced solid tumors | 86 | Not yet recruiting | No results posted |
CRC: colorectal cancer; mCRC: metastatic CRC; MSS: microsatellite stability; MSI: microsatellite instability; MSI-H: microsatellite instability-high; dMMR: deficient mismatch repair; RCC: renal cell carcinoma; NSCLC: non-small cell lung cancer; ORR: objective response rate; PFS: progression free survival; OS: overall survival; PR: partial response; RECIST: response evaluation criteria in solid tumors.
MSI is extremely rare in breast cancer, showing a frequency of 0.04–3% [158,159]. However, MSI was observed in triple negative breast cancer (TNBC), with a high frequency accounting for between approximately 0.2% and 18,6% [160,161,162]. The role of MSI in BC is still uncertain. Some studies have shown that MSI does not have a significant impact on patient survival [149]. Conversely, other studies have reported a positive prognostic value of MSI in TNBC [160]. A significant correlation was observed between MSI and the negative expression of estrogen and progesterone receptors, indicating a possible relationship between genetic changes in microsatellite regions and hormonal deregulation in the progression of BC [163]. In BC, MSI showed a correlation with the advanced clinical stage, the tumor size and the high grade [164]. The KEYNOTE-028 and KEYNOTE-012 studies demonstrated the efficacy of pembrolizumab in BC previously treated with CDK4/6 inhibitors. [158,165]. The pancreatic ductal adenocarcinoma (PDAC) showed MSI in about 1–2% [166,167]. PDACs carrying MSI are generally associated with LS [168]. LS-associated pancreatic tumors, ranging from 0.5% to 3.9%, are generally associated with medullary histology showing prominent lymphocyte infiltration [167,169]. PDAC is an aggressive type of human cancer; moreover, most patients are resistant to chemotherapy. Thus, immunotherapy could be a possible therapeutic option [170]. Previous studies showed that pMMR PDCA patients treated with conventional chemotherapy have a survival advantage compared to dMMR patients, suggesting that MSI can be considered a potential predictor of treatment sensitivity [171,172,173]. KEYNOTE-016 evaluated the efficacy of pembrolizumab in eight cases of MSI PDAC, showing a complete response in two patients and a partial response in three patients [27,174]. These preliminary results suggested that anti-PD-L1 antibody treatment could be effective in patients with MSI PDAC. MSI was reported in approximately 10–15% of thyroid carcinomas (TC) [175,176,177]; moreover, a correlation between MSI and clinical-pathological features was observed. Particularly, MSI TCs were generally papillary or anaplastic and poorly differentiated histotypes. A recent study showed that MSI TC patients have a good prognosis compared to MSS TC [177]. Although several clinical trials are being evaluated for the efficacy of pembrolizumab, nivolumab and atezolizumab in MSI TC, until now no immunological therapy has been approved for MSI TC [178,179]. The ICI targeting the PD-1/PD-L1 pathway may be a treatment option for MSI TC; however, further studies are needed [177]. The frequency of MSI brain tumors is very low, ranging from 2% to 3% of cases [27,116]. Brain tumors are rarely associated with LS (0.5–3.7% of all cases). No information is available regarding the specific molecular or phenotypic characteristics of LS-related brain tumors [167]. The MSI phenotype was observed in about 25% of glioblastomas (GBM) [180]. MSI GBM showed a low level of PD-L1 expression, suggesting it is not a good predictive marker for ICI targeting the PD1/PD-L1 pathway [181]. MSI has been shown to occur in malignant melanomas, with a frequency of between 2 and 30% [182,183,184]. Kubecek et al. suggested that MSI might be a predictive marker in malignant melanoma [185]. Although the KEYNOTE-016 study showed the efficacy of pembrolizumab in MSI melanomas that have progressed after previous treatment, the treatment for MSI melanoma is not yet used in clinical practice [27].
MSI has been identified in approximately 1–28% of urothelial carcinomas (UC) [186,187,188]. The upper tract urothelial carcinomas (UTUCs) also occur in about 5% of patients with LS [189]. The heterodimer MSH2/MSH6 is frequently lost in the UTUC in about 50–86% of cases [186,190,191]. Several studies reported that UTUCs MSI showed unique morphological features compared to MSS UTUC, including increased intratumoral lymphocytes, the lack of nuclear pleomorphism and the presence of pushing edges [167,190,192]. The KEYNOTE-158 study demonstrated the benefit of pembrolizumab treatment in patients carrying MSI-H/dMMR including UCs [174]. The prostate cancers (PCs) showed MSI status in 1,2–12% of cases. MSI PCs represent a clinically aggressive phenotype [193]. The germline alterations in the MMR genes are less common in localized PC, suggesting an increase in mutations in patients with metastatic spread [194]. In the KEYNOTE-016 study, MSI PCs responded better to ICI [27]. Although MSI can be considered a biomarker capable of responding to immunotherapy in prostate cancer, ICIs are not yet used in clinical practice for the treatment of metastatic PCs [195].
7. Conclusions
The MMR system is a key repair mechanism for maintaining sequence fidelity and stability. The inactivation of the MMR system due to the germinal, somatic or epigenetic prevents error correction and, thus, promotes microsatellite instability. MSI has been detected in various cancers with a prognostic and/or predictive role; in particular, it is clinically important for predicting a response to immunotherapy. MSI represents the FDA’s first cancer type-agnostic biomarker approved for selection to the treatment with pembrolizumab of patients with any advanced solid cancer, regardless of the histology. The results reported underline the pivotal role of MSI in the choice of drug treatment in patients, especially in the cases of those who no longer respond to chemotherapy. To date, MSI detection aimed at selecting patients for ICI treatment could be performed through both IHC and PCR/NGS. Although the two tests are equated in clinical practice for the therapeutic choice, several controversial results have been reported. In this view, the optimization of MSI detection in clinical practice is currently needed in order to implement an adequate selection of patients eligible for ICI treatment of dMMR tumors. A feasible diagnostic algorithm for MSI testing could include IHC as prescreening requiring an additional molecular test for cases with an equivocal IHC of one or more MMR proteins, and also to confirm dMMR cases in order to exclude false IHC results.
Author Contributions
Conceptualization, M.A. (Marina Accardo) and F.Z.M.; writing—original draft preparation, M.A. (Martina Amato), M.A. (Marina Accardo), F.Z.M. and R.F.; review and editing, R.A., F.C., M.A. (Marina Accardo), G.V., A.S., G.F., M.B., S.P. and M.C.; supervision, F.Z.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li K., Luo H., Huang L., Luo H., Zhu X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020;20:16. doi: 10.1186/s12935-019-1091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hause R.J., Pritchard C.C., Shendure J., Salipante S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 3.Kawka M., Parada R., Jaszczak K., Horbańczuk J.O. The use of microsatellite polymorphism in genetic mapping of the ostrich (Struthio camelus) Mol. Biol. Rep. 2012;39:3369–3374. doi: 10.1007/s11033-011-1107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastos-Rodrigues L., Pimenta J.R., Pena S.D.J. The Genetic Structure of Human Populations Studied Through Short Inser-tion-Deletion Polymorphisms. Ann. Hum. Genet. 2006;70:658–665. doi: 10.1111/j.1469-1809.2006.00287.x. [DOI] [PubMed] [Google Scholar]
- 5.Sturzeneker R., Bevilacqua R.A., Haddad L.A., Simpson A.J., Pena S.D. Microsatellite instability in tumors as a model to study the process of microsatellite mutations. Hum. Mol. Genet. 2000;9:347–352. doi: 10.1093/hmg/9.3.347. [DOI] [PubMed] [Google Scholar]
- 6.Eso Y., Shimizu T., Takeda H., Takai A., Marusawa H. Microsatellite instability and immune checkpoint inhibitors: Toward pre-cision medicine against gastrointestinal and hepatobiliary cancers. J. Gastroenterol. 2020;55:15–26. doi: 10.1007/s00535-019-01620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 8.Bonneville R., Krook M.A., Chen H.Z., Smith A., Samorodnitsky E., Wing M.R., Reeser J.W., Roychowdhury S. Detection of Mi-crosatellite Instability Biomarkers via Next-Generation Sequencing. Methods Mol. Biol. 2020;2055:119–132. doi: 10.1007/978-1-4939-9773-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lochhead P., Kuchiba A., Imamura Y., Liao X., Yamauchi M., Nishihara R., Qian Z.R., Morikawa T., Shen J., Meyerhardt J.A., et al. Microsatellite Instability and BRAF Mutation Testing in Colorectal Cancer Prognostication. J. Natl. Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligtenberg M.J., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J., et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 11.Edwards R.A., Witherspoon M., Wang K., Afrasiabi K., Pham T., Birnbaumer L., Lipkin S.M. Epigenetic repression of DNA mis-match repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto H., Adachi Y., Taniguchi H., Kunimoto H., Nosho K., Suzuki H., Shinomura Y. Interrelationship between microsatel-lite instability and microRNA in gastrointestinal cancer. World J. Gastroenterol. 2012;18:2745–2755. doi: 10.3748/wjg.v18.i22.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai Y., Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 2003;20:2123–2131. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- 14.Gologan A., Sepulveda A.R. Microsatellite Instability and DNA Mismatch Repair Deficiency Testing in Hereditary and Sporadic Gastrointestinal Cancers. Clin. Lab. Med. 2005;25:179–196. doi: 10.1016/j.cll.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Yurgelun M.B., Kastrinos F. Tumor Testing for Microsatellite Instability to Identify Lynch Syndrome: New Insights into an Old Diagnostic Strategy. J. Clin. Oncol. 2019;37:263–265. doi: 10.1200/JCO.18.01664. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez-Valentin M., Sampson J.R., Seppälä T.T., Ten Broeke S.W., Plazzer J.P., Nakken S., Engel C., Aretz S., Jenkins M.A., Sunde L., et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the Prospective Lynch Syndrome Database. Genet. Med. 2020;22:15–25. doi: 10.1038/s41436-019-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bucksch K., Zachariae S., Aretz S., Büttner R., Holinski-Feder E., Holzapfel S., Hüneburg R., Kloor M., von Doeberitz M.K., Morak M., et al. Cancer risks in Lynch syndrome, Lynch-like syndrome, and familial colorectal cancer type X: A prospective cohort study. BMC Cancer. 2020;20:460. doi: 10.1186/s12885-020-06926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim T.-M., Laird P.W., Park P.J. The Landscape of Microsatellite Instability in Colorectal and Endometrial Cancer Genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oda S., Maehara Y., Ikeda Y., Oki E., Egashira A., Okamura Y., Takahashi I., Kakeji Y., Sumiyoshi Y., Miyashita K., et al. Two modes of microsatellite insta-bility in human cancer: Differential connection of defective DNA mismatch repair to dinucleotide repeat instability. Nucleic Acids Res. 2005;33:1628–1636. doi: 10.1093/nar/gki303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibodeau S.N., Bren G., Schaid D. Microsatellite Instability in Cancer of the Proximal Colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 21.De la Chapelle A., Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J. Clin. Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard C.C., Morrissey C., Kumar A., Zhang X., Smith C., Coleman I., Salipante S.J., Milbank J., Yu M., Grady W.M., et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat. Commun. 2014;5:4988. doi: 10.1038/ncomms5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley J.C., Lin M.-T., Le D.T., Eshleman J.R. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin. Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi Y., Kondo K., Hirose T., Nakagawa H., Tsuyuguchi M., Hashimoto M., Sano T., Ochiai A., Monden Y. Microsatellite instability and protein expression of the DNA mismatch repair gene, hMLH1, of lung cancer in chromate-exposed workers. Mol. Carcinog. 2005;42:150–158. doi: 10.1002/mc.20073. [DOI] [PubMed] [Google Scholar]
- 25.Alvino E., Marra G., Pagani E., Falcinelli S., Pepponi R., Perrera C., Haider R., Castiglia D., Ferranti G., Bonmassar E., et al. High-Frequency Microsatellite Instability is Associated with Defective DNA Mismatch Repair in Human Melanoma. J. Investig. Dermatol. 2002;118:79–86. doi: 10.1046/j.0022-202x.2001.01611.x. [DOI] [PubMed] [Google Scholar]
- 26.Kassem H.S., Varley J.M., Hamam S.M., Margison G.P. Immunohistochemical analysis of expression and allelotype of mismatch repair genes (hMLH1 and hMSH2) in bladder cancer. Br. J. Cancer. 2001;84:321–328. doi: 10.1054/bjoc.2000.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le D.T., Durham J.N., Smith K.N., Wang H., Bartlett B.R., Aulakh L.K., Lu S., Kemberling H., Wilt C., Luber B.S., et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jover R., Zapater P., Castells A., Llor X., Andreu M., Cubiella J., Balaguer F., Sempere L., Xicola R.M., Bujanda L., et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur. J. Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Nikanjam M., Arguello D., Gatalica Z., Swensen J., Barkauskas D.A., Kurzrock R. Relationship between protein biomarkers of chemotherapy response and microsatellite status, tumor mutational burden and PD-L1 expression in cancer patients. Int. J. Cancer. 2019;146:3087–3097. doi: 10.1002/ijc.32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drescher K.M., Sharma P., Watson P., Gatalica Z., Thibodeau S.N., Lynch H.T. Lymphocyte recruitment into the tumor site is altered in patients with MSI-H colon cancer. Fam. Cancer. 2009;8:231–239. doi: 10.1007/s10689-009-9233-0. [DOI] [PubMed] [Google Scholar]
- 31.Llosa N.J., Cruise M., Tam A., Wicks E.C., Hechenbleikner E.M., Taube J.M., Blosser R.L., Fan H., Wang H., Luber B.S., et al. The vigorous immune microenvi-ronment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C., Patel K., Singhi A.D., Ren B., Zhu B., Shaikh F., Sun W. Programmed Death-Ligand 1 Expression Is Common in Gastric Cancer Associated with Epstein-Barr Virus or Microsatellite Instability. Am. J. Surg. Pathol. 2016;40:1496–1506. doi: 10.1097/PAS.0000000000000698. [DOI] [PubMed] [Google Scholar]
- 33.Bai W., Ma J., Liu Y., Liang J., Wu Y., Yang X., Xu E., Li Y., Xi Y. Screening of MSI detection loci and their heterogeneity in East Asian colorectal cancer patients. Cancer Med. 2019;8:2157–2166. doi: 10.1002/cam4.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Z., Yang B., Liao Z. Biomarkers in Immunotherapy-Based Precision Treatments of Digestive System Tumors. Front. Oncol. 2021;11:650481. doi: 10.3389/fonc.2021.650481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marginean E.C., Melosky B. Is There a Role for Programmed Death Ligand-1 Testing and Immunotherapy in Colorectal Can-cer With Microsatellite Instability? Part I-Colorectal Cancer: Microsatellite Instability, Testing, and Clinical Implications. Arch. Pathol. Lab. Med. 2018;142:17–25. doi: 10.5858/arpa.2017-0040-RA. [DOI] [PubMed] [Google Scholar]
- 36.Pearlman R., Markow M., Knight D., Chen W., Arnold C.A., Pritchard C.C., Hampel H., Frankel W.L. Two-stain immunohisto-chemical screening for Lynch syndrome in colorectal cancer may fail to detect mismatch repair deficiency. Mod. Pathol. 2018;31:1891–1900. doi: 10.1038/s41379-018-0058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W., Swanson B.J., Frankel W.L. Molecular genetics of microsatellite-unstable colorectal cancer for pathologists. Diagn. Pathol. 2017;12:24. doi: 10.1186/s13000-017-0613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigau V., Sebbagh N., Olschwang S., Paraf F., Mourra N., Parc Y., Flejou J.F. Microsatellite instability in colorectal carcinoma. The comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 [correction of hMLH6] im-munostaining. Arch. Pathol. Lab. Med. 2003;127:694–700. doi: 10.5858/2003-127-694-MIICC. [DOI] [PubMed] [Google Scholar]
- 39.Beamer L., Grant M.L., Espenschied C., Blazer K.R., Hampel H.L., Weitzel J.N., Macdonald D.J. Reflex Immunohistochemistry and Microsatellite Instability Testing of Colorectal Tumors for Lynch Syndrome Among US Cancer Programs and Follow-Up of Abnormal Results. J. Clin. Oncol. 2012;30:1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hampel H. Point: Justification for Lynch Syndrome Screening Among All Patients with Newly Diagnosed Colorectal Cancer. J. Natl. Compr. Cancer Netw. 2010;8:597–601. doi: 10.6004/jnccn.2010.0044. [DOI] [PubMed] [Google Scholar]
- 41.Berardinelli G.N., Scapulatempo-Neto C., Durães R., de Oliveira M.A., Guimarães D., Reis R.M. Advantage of HSP110 (T17) marker inclusion for microsatellite instability (MSI) detection in colorectal cancer patients. Oncotarget. 2018;9:28691–28701. doi: 10.18632/oncotarget.25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bacher J.W., Sievers C.K., Albrecht D.M., Grimes I.C., Weiss J., Matkowskyj K.A., Agni R.M., Vyazunova I., Clipson L., Storts U.R., et al. Improved Detection of Microsatellite Instability in Early Colorectal Lesions. PLoS ONE. 2015;10:e0132727. doi: 10.1371/journal.pone.0132727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy A.J., Capo-Chichi J.M., Spence T., Grenier S., Stockley T., Kamel-Reid S., Serra S., Sabatini P., Chetty R. Heterogenous loss of mismatch repair (MMR) protein expression: A challenge for immunohistochemical interpretation and microsatellite in-stability (MSI) evaluation. J. Pathol. Clin. Res. 2019;5:115–129. doi: 10.1002/cjp2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathiak M., Warneke V.S., Behrens H.M., Haag J., Böger C., Krüger S., Röcken C. Clinicopathologic Characteristics of Microsatel-lite Instable Gastric Carcinomas Revisited: Urgent Need for Standardization. Appl. Immunohistochem. Mol. Morphol. 2017;25:12–24. doi: 10.1097/PAI.0000000000000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartley A.N., Luthra R., Saraiya D.S., Urbauer D.L., Broaddus R.R. Identification of cancer patients with Lynch syndrome: Clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev. Res. 2012;5:320–327. doi: 10.1158/1940-6207.CAPR-11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suraweera N., Duval A., Reperant M., Vaury C., Furlan D., Leroy K., Seruca R., Iacopetta B., Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 47.Campanella N.C., Berardinelli G.N., Scapulatempo-Neto C., Viana D., Palmero E.I., Pereira R., Reis R.M. Optimization of a penta-plex panel for MSI analysis without control DNA in a Brazilian population: Correlation with ancestry markers. Eur. J. Hum. Genet. 2014;22:875–880. doi: 10.1038/ejhg.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umar A., Risinger J.I., Hawk E.T., Barrett J.C. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat. Cancer. 2004;4:153–158. doi: 10.1038/nrc1278. [DOI] [PubMed] [Google Scholar]
- 49.Murphy K.M., Zhang S., Geiger T., Hafez M.J., Bacher J., Berg K.D., Eshleman J.R. Comparison of the Microsatellite Instability Analysis System and the Bethesda Panel for the Determination of Microsatellite Instability in Colorectal Cancers. J. Mol. Diagn. 2006;8:305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheah P.L., Li J., Looi L.M., Koh C.C., Lau T.P., Chang S.W., Teoh K.H., Mun K.S., Nazarina A.R. Screening for microsatellite instability in colorectal carcinoma: Practical utility of immunohistochemistry and PCR with fragment analysis in a diagnostic histopathology setting. Malays. J. Pathol. 2019;41:91–100. [PubMed] [Google Scholar]
- 51.Cho J., Kang S.Y., Kim K.-M. MMR protein immunohistochemistry and microsatellite instability in gastric cancers. Pathology. 2018;51:110–113. doi: 10.1016/j.pathol.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 52.Haron N.H., Mohamad Hanif E.A., Abdul Manaf M.R., Yaakub J.A., Harun R., Mohamed R., Mohamed Rose I. Microsatellite In-stability and Altered Expressions of MLH1 and MSH2 in Gastric Cancer. Asian Pac. J. Cancer Prev. 2019;20:509–517. doi: 10.31557/APJCP.2019.20.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salipante S.J., Scroggins S.M., Hampel H.L., Turner E.H., Pritchard C.C. Microsatellite instability detection by next generation se-quencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 54.Lu Y., Soong T.D., Elemento O. A Novel Approach for Characterizing Microsatellite Instability in Cancer Cells. PLoS ONE. 2013;8:e63056. doi: 10.1371/journal.pone.0063056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D., Wendl M.C., Ding L. MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2013;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kautto E.A., Bonneville R., Miya J., Yu L., Krook M.A., Reeser J.W., Roychowdhury S. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2016;8:7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L., et al. ColoSeq Provides Comprehensive Lynch and Polyposis Syndrome Mutational Analysis Using Massively Parallel Sequencing. J. Mol. Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu L., Huang Y., Fang X., Liu C., Deng W., Zhong C., Xu J., Xu D., Yuan Y. A Novel and Reliable Method to Detect Microsatel-lite Instability in Colorectal Cancer by Next-Generation Sequencing. J. Mol. Diagn. 2018;20:225–231. doi: 10.1016/j.jmoldx.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Salem M.E., Battaglin F., Goldberg R.M., Puccini A., Shields A.F., Arguello D., Korn W.M., Marshall J.L., Grothey A., Lenz H.J. Molecular Analyses of Left- and Right-Sided Tumors in Adolescents and Young Adults with Colorectal Cancer. Oncologist. 2020;25:404–413. doi: 10.1634/theoncologist.2019-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zito Marino F., Amato M., Ronchi A., Panarese I., Ferraraccio F., De Vita F., Tirino G., Martinelli E., Troiani T., Facchini G., et al. Microsatellite Status Detection in Gastrointestinal Cancers: PCR/NGS Is Mandatory in Negative/Patchy MMR Immunohistochemistry. Cancers. 2022;14:2204. doi: 10.3390/cancers14092204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The Cancer Genome Atlas (TCGA) Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goel A., Nagasaka T., Arnold C.N., Inoue T., Hamilton C., Niedzwiecki D., Compton C., Mayer R.J., Goldberg R., Bertagnolli M.M., et al. The CpG Island Methylator Phenotype and Chromosomal Instability Are Inversely Correlated in Sporadic Colorectal Cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 63.Li L.S., Kim N.-G., Kim S.H., Park C., Kim H., Kang H.J., Koh K.H., Kim S.N., Kim W.H., Kim H. Chromosomal Imbalances in the Colorectal Carcinomas with Microsatellite Instability. Am. J. Pathol. 2003;163:1429–1436. doi: 10.1016/S0002-9440(10)63500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang W., Cai M.-Y., Li S.-Y., Bei J.-X., Wang F., Hampel H., Ling Y.-H., Frayling I.M., Sinicrope F.A., Rodriguez-Bigas M.A., et al. Universal screening for Lynch syndrome in a large consecutive cohort of Chinese colorectal cancer patients: High prevalence and unique molecular features. Int. J. Cancer. 2018;144:2161–2168. doi: 10.1002/ijc.32044. [DOI] [PubMed] [Google Scholar]
- 65.Li D., Hoodfar E., Jiang S.F., Udaltsova N., Pham N.P., Jodesty Y., Armstrong M.A., Hung Y.Y., Baker R.J., Postlethwaite D., et al. Comparison of Universal Versus Age-Restricted Screening of Colorectal Tumors for Lynch Syndrome Using Mismatch Repair Immunohistochemistry: A Cohort Study. Ann. Intern. Med. 2019;171:19–26. doi: 10.7326/M18-3316. [DOI] [PubMed] [Google Scholar]
- 66.Nakayama Y., Iijima T., Inokuchi T., Kojika E., Takao M., Takao A., Koizumi K., Horiguchi S.I., Hishima T., Yamaguchi T. Clinicopathological features of sporadic MSI colorectal cancer and Lynch syndrome: A single-center retrospective cohort study. Int. J. Clin. Oncol. 2021;26:1881–1889. doi: 10.1007/s10147-021-01968-y. [DOI] [PubMed] [Google Scholar]
- 67.Nojadeh J.N., Behrouz Sharif S., Sakhinia E. Microsatellite instability in colorectal cancer. EXCLI J. 2018;17:159–168. doi: 10.17179/excli2017-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran B., Kopetz S., Tie J., Gibbs P., Jiang Z.-Q., Lieu C.H., Agarwal A., Maru D.M., Sieber O., Desai J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–4632. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ladas I., Yu F., Leong K.W., Fitarelli-Kiehl M., Song C., Ashtaputre R., Kulke M., Mamon H., Makrigiorgos G.M. Enhanced detec-tion of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic Acids Res. 2018;46:e74. doi: 10.1093/nar/gky251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kloor M., Huth C., Voigt A.Y., Benner A., Schirmacher P., von Knebel Doeberitz M., Bläker H. Prevalence of mismatch re-pair-deficient crypt foci in Lynch syndrome: A pathological study. Lancet Oncol. 2012;13:598–606. doi: 10.1016/S1470-2045(12)70109-2. [DOI] [PubMed] [Google Scholar]
- 71.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pino M.S., Mino-Kenudson M., Wildemore B.M., Ganguly A., Batten J., Sperduti I., Iafrate A.J., Chung D.C. Deficient DNA mis-match repair is common in Lynch syndrome-associated colorectal adenomas. J. Mol. Diagn. 2009;11:238–247. doi: 10.2353/jmoldx.2009.080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venderbosch S., Nagtegaal I.D., Maughan T.S., Smith C.G., Cheadle J.P., Fisher D., Kaplan R., Quirke P., Seymour M.T., Richman S.D., et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: A pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin. Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith C.G., Fisher D., Claes B., Maughan T.S., Idziaszczyk S., Peuteman G., Harris R., James M.D., Meade A., Jasani B., et al. Somatic Profiling of the Epidermal Growth Factor Receptor Pathway in Tumors from Patients with Advanced Colorectal Cancer Treated with Chemotherapy ± Cetuximab. Clin. Cancer Res. 2013;19:4104–4113. doi: 10.1158/1078-0432.CCR-12-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le D.T., Kim T.W., Van Cutsem E., Geva R., Jäger D., Hara H., Burge M., O’Neil B., Kavan P., Yoshino T., et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020;38:11–19. doi: 10.1200/JCO.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X., Wu T., Cai X., Dong J., Xia C., Zhou Y., Ding R., Yang R., Tan J., Zhang L., et al. Neoadjuvant Immunotherapy for MSI-H/dMMR Locally Advanced Colorectal Cancer: New Strategies and Unveiled Opportunities. Front. Immunol. 2022;13:795972. doi: 10.3389/fimmu.2022.795972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andre T., Amonkar M., Norquist J.M., Shiu K.-K., Kim T.W., Jensen B.V., Jensen L.H., Punt C.J.A., Smith D., Garcia-Carbonero R., et al. Health-related quality of life in patients with microsatellite instability-high or mismatch repair deficient metastatic colorectal cancer treated with first-line pembrolizumab versus chemotherapy (KEYNOTE-177): An open-label, randomised, phase 3 trial. Lancet Oncol. 2021;22:665–677. doi: 10.1016/S1470-2045(21)00064-4. [DOI] [PubMed] [Google Scholar]
- 78.Overman M.J., Lonardi S., Wong K.Y.M., Lenz H.J., Gelsomino F., Aglietta M., Morse M.A., Van Cutsem E., McDermott R., Hill A., et al. Durable Clinical Benefit with Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 79.Lenz H.-J., Van Cutsem E., Limon M.L., Wong K.Y.M., Hendlisz A., Aglietta M., García-Alfonso P., Neyns B., Luppi G., Cardin D.B., et al. First-Line Nivolumab Plus Low-Dose Ipilimumab for Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: The Phase II CheckMate 142 Study. J. Clin. Oncol. 2022;40:161–170. doi: 10.1200/JCO.21.01015. [DOI] [PubMed] [Google Scholar]
- 80.Hatch S.B., Lightfoot H.M., Jr., Garwacki C.P., Moore D.T., Calvo B.F., Woosley J.T., Sciarrotta J., Funkhouser W.K., Farber R.A. Microsatellite instability testing in colorectal carcinoma: Choice of markers affects sensitivity of detection of mismatch re-pair-deficient tumors. Clin. Cancer Res. 2005;11:2180–2187. doi: 10.1158/1078-0432.CCR-04-0234. [DOI] [PubMed] [Google Scholar]
- 81.Hissong E., Crowe E.P., Yantiss R.K., Chen Y.-T. Assessing colorectal cancer mismatch repair status in the modern era: A survey of current practices and re-evaluation of the role of microsatellite instability testing. Mod. Pathol. 2018;31:1756–1766. doi: 10.1038/s41379-018-0094-7. [DOI] [PubMed] [Google Scholar]
- 82.Yuan L., Chi Y., Chen W., Chen X., Wei P., Sheng W., Zhou X., Shi D. Immunohistochemistry and microsatellite instability analy-sis in molecular subtyping of colorectal carcinoma based on mismatch repair competency. Int. J. Clin. Exp. Med. 2015;8:20988–21000. [PMC free article] [PubMed] [Google Scholar]
- 83.Dedeurwaerdere F., Claes K.B., Van Dorpe J., Rottiers I., Van der Meulen J., Breyne J., Swaerts K., Martens G. Comparison of mi-crosatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021;11:12880. doi: 10.1038/s41598-021-91974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loughrey M.B., McGrath J., Coleman H.G., Bankhead P., Maxwell P., McGready C., Bingham V., Humphries M.P., Craig S.G., McQuaid S., et al. Identifying mismatch repair-deficient colon cancer: Near-perfect concordance between immunohistochemistry and microsatellite instability testing in a large, population-based series. Histopathology. 2021;78:401–413. doi: 10.1111/his.14233. [DOI] [PubMed] [Google Scholar]
- 85.Xiao J., Li W., Huang Y., Huang M., Li S., Zhai X., Zhao J., Gao C., Xie W., Qin H., et al. A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer. BMC Cancer. 2021;21:282. doi: 10.1186/s12885-021-07942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilson P., Levy J., Rouyer M., Demange J., Husson M., Bonnet C., Salleron J., Leroux A., Merlin J.L., Harlé A. Evaluation of 3 mo-lecular-based assays for microsatellite instability detection in formalin-fixed tissues of patients with endometrial and colorectal cancers. Sci. Rep. 2020;10:16386. doi: 10.1038/s41598-020-73421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jang M., Kwon Y., Kim H., Kim H., Min B.S., Park Y., Kim T.I., Hong S.P., Kim W.K. Microsatellite instability test using peptide nu-cleic acid probe-mediated melting point analysis: A comparison study. BMC Cancer. 2018;18:1218. doi: 10.1186/s12885-018-5127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang W. TCGA divides gastric cancer into four molecular subtypes: Implications for individualized therapeutics. Chin. J. Cancer. 2014;33:469–470. doi: 10.5732/cjc.014.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang N., Wu Y., Jin M., Jia Z., Wang Y., Cao D., Qin L., Wang X., Zheng M., Cao X., et al. Microsatellite instability and Ep-stein-Barr virus combined with PD-L1 could serve as a potential strategy for predicting the prognosis and efficacy of postop-erative chemotherapy in gastric cancer. PeerJ. 2021;9:e11481. doi: 10.7717/peerj.11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cristescu R., Lee J., Nebozhyn M., Kim K.-M., Ting J.C., Wong S.S., Liu J., Yue Y.G., Wang J., Yu K., et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 91.Guan W.L., Ma Y., Cui Y.H., Liu T.S., Zhang Y.Q., Zhou Z.W., Xu J.Y., Yang L.Q., Li J.Y., Sun Y.T., et al. The Impact of Mismatch Repair Status on Prognosis of Patients with Gastric Cancer: A Multicenter Analysis. Front. Oncol. 2021;11:712760. doi: 10.3389/fonc.2021.712760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sugimoto R., Sugai T., Habano W., Endoh M., Eizuka M., Yamamoto E., Uesugi N., Ishida K., Kawasaki T., Matsumoto T., et al. Clinicopathological and molecular alterations in early gastric cancers with the microsatellite instability-high phenotype. Int. J. Cancer. 2015;138:1689–1697. doi: 10.1002/ijc.29916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ottini L., Falchetti M., Lupi R., Rizzolo P., Agnese V., Colucci G., Bazan V., Russo A. Patterns of genomic instability in gastric cancer: Clinical implications and perspectives. Ann. Oncol. 2006;17:vii97–vii102. doi: 10.1093/annonc/mdl960. [DOI] [PubMed] [Google Scholar]
- 94.Corso G., Velho S., Paredes J., Pedrazzani C., Martins D., Milanezi F., Pascale V., Vindigni C., Pinheiro H., Leite M., et al. Oncogenic mutations in gastric cancer with microsatellite instability. Eur. J. Cancer. 2011;47:443–451. doi: 10.1016/j.ejca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 95.Hudler P. Genetic Aspects of Gastric Cancer Instability. Sci. World J. 2012;2012:761909. doi: 10.1100/2012/761909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao W., Chan T.L., Chu K.-M., Chan A.S., Stratton M.R., Yuen S.T., Leung S.Y. Mutations ofBRAF andKRAS in gastric cancer and their association with microsatellite instability. Int. J. Cancer. 2003;108:167–169. doi: 10.1002/ijc.11553. [DOI] [PubMed] [Google Scholar]
- 97.Huang H., Wang Z., Li Y., Zhao Q., Niu Z. Amplification of the human epidermal growth factor receptor 2 (HER2) gene is as-sociated with a microsatellite stable status in Chinese gastric cancer patients. J. Gastrointest. Oncol. 2021;12:377–387. doi: 10.21037/jgo-21-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Polom K., Marano L., Marrelli D., DE Luca R., Roviello G., Savelli V., Tan P. Meta-analysis of microsatellite instability in relation to clinicopathological characteristics and overall survival in gastric cancer. Br. J. Surg. 2017;105:159–167. doi: 10.1002/bjs.10663. [DOI] [PubMed] [Google Scholar]
- 99.Cai L., Sun Y., Wang K., Guan W., Yue J., Li J., Wang R., Wang L. The Better Survival of MSI Subtype Is Associated With the Oxidative Stress Related Pathways in Gastric Cancer. Front. Oncol. 2020;10:1269. doi: 10.3389/fonc.2020.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Q., Wang L., Ni S., Tan C., Cai X., Huang D., Sheng W. Clinicopathological features and prognostic value of mismatch repair protein deficiency in gastric cancer. Int. J. Clin. Exp. Pathol. 2018;11:2579–2587. [PMC free article] [PubMed] [Google Scholar]
- 101.An J.Y., Kim H., Cheong J.-H., Hyung W.J., Kim H., Noh S.H. Microsatellite instability in sporadic gastric cancer: Its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int. J. Cancer. 2011;131:505–511. doi: 10.1002/ijc.26399. [DOI] [PubMed] [Google Scholar]
- 102.Smyth E.C., Wotherspoon A., Peckitt C., Gonzalez D., Hulkki-Wilson S., Eltahir Z., Fassan M., Rugge M., Valeri N., Okines A., et al. Mismatch Repair Deficiency, Microsatellite Instability, and Survival: An Exploratory Analysis of the Medical Research Council Adjuvant Gastric Infusional Chemotherapy (MAGIC) Trial. JAMA Oncol. 2017;3:1197–1203. doi: 10.1001/jamaoncol.2016.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Janjigian Y.Y., Sanchez-Vega F., Jonsson P., Chatila W.K., Hechtman J.F., Ku G.Y., Riches J.C., Tuvy Y., Kundra R., Bouvier N., et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer Discov. 2018;8:49–58. doi: 10.1158/2159-8290.CD-17-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chao J., Fuchs C.S., Shitara K., Tabernero J., Muro K., Van Cutsem E., Bang Y.J., De Vita F., Landers G., Yen C.J., et al. Assessment of Pembrolizumab Therapy for the Treatment of Microsatellite Instability-High Gastric or Gastroesophageal Junction Cancer Among Patients in the KEYNOTE-059, KEYNOTE-061, and KEYNOTE-062 Clinical Trials. JAMA Oncol. 2021;7:895–902. doi: 10.1001/jamaoncol.2021.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cohen R., Pudlarz T., Garcia-Larnicol M.L., Vernerey D., Dray X., Clavel L., Jary M., Piessen G., Zaanan A., Aparicio T., et al. Localized MSI/dMMR gastric cancer patients, perioperative immunotherapy instead of chemotherapy: The GERCOR NE-ONIPIGA phase II study is opened to recruitment. Bull Cancer. 2020;107:438–446. doi: 10.1016/j.bulcan.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 106.Raimondi A., Palermo F., Prisciandaro M., Aglietta M., Antonuzzo L., Aprile G., Berardi R., Cardellino G.G., De Manzoni G., De Vita F., et al. TremelImumab and Durvalumab Combination for the Non-OperatIve Management (NOM) of Microsatellite InstabiliTY (MSI)-High Resectable Gastric or Gastroesophageal Junction Cancer: The Multicentre, Single-Arm, Multi-Cohort, Phase II INFINITY Study. Cancers. 2021;13:2839. doi: 10.3390/cancers13112839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park J., Shin S., Yoo H.M., Lee S.W., Kim J.G. Evaluation of the Three Customized MSI Panels to Improve the Detection of Mi-crosatellite Instability in Gastric Cancer. Clin Lab. 2017;63:705–716. doi: 10.7754/Clin.Lab.2016.161029. [DOI] [PubMed] [Google Scholar]
- 108.Pécriaux A., Favre L., Calderaro J., Charpy C., Derman J., Pujals A. Detection of microsatellite instability in a panel of solid tu-mours with the Idylla MSI Test using extracted DNA. J. Clin Pathol. 2021;74:36–42. doi: 10.1136/jclinpath-2020-206581. [DOI] [PubMed] [Google Scholar]
- 109.Farmkiss L., Hopkins I., Jones M. Idylla microsatellite instability assay versus mismatch repair immunohistochemistry: A retro-spective comparison in gastric adenocarcinoma. J. Clin Pathol. 2021;74:604–607. doi: 10.1136/jclinpath-2020-207033. [DOI] [PubMed] [Google Scholar]
- 110.Aysal A., Karnezis A., Medhi I., Grenert J.P., Zaloudek C.J., Rabban J.T. Ovarian endometrioid adenocarcinoma: Incidence and clinical significance of the morphologic and immunohistochemical markers of mismatch repair protein defects and tumor mi-crosatellite instability. Am. J. Surg. Pathol. 2012;36:163–172. doi: 10.1097/PAS.0b013e31823bc434. [DOI] [PubMed] [Google Scholar]
- 111.Jensen K.C., Mariappan M.R., Putcha G.V., Husain A., Chun N., Ford J.M., Schrijver I., Longacre T.A. Microsatellite Instability and Mismatch Repair Protein Defects in Ovarian Epithelial Neoplasms in Patients 50 Years of Age and Younger. Am. J. Surg. Pathol. 2008;32:1029–1037. doi: 10.1097/PAS.0b013e31816380c4. [DOI] [PubMed] [Google Scholar]
- 112.Murphy M.A., Wentzensen N. Frequency of mismatch repair deficiency in ovarian cancer: A systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int. J. Cancer. 2010;129:1914–1922. doi: 10.1002/ijc.25835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Howitt B.E., Strickland K.C., Sholl L.M., Rodig S., Ritterhouse L.L., Chowdhury D., D’Andrea A.D., Matulonis U.A., Konstantinopoulos P.A. Clear cell ovarian cancers with microsatellite instability: A unique subset of ovarian cancers with increased tu-mor-infiltrating lymphocytes and PD-1/PD-L1 expression. Oncoimmunology. 2017;6:e1277308. doi: 10.1080/2162402X.2016.1277308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shilpa V., Bhagat R., Premalata C.S., Pallavi V.R., Krishnamoorthy L. Microsatellite instability, promoter methylation and protein expression of the DNA mismatch repair genes in epithelial ovarian cancer. Genomics. 2014;104:257–263. doi: 10.1016/j.ygeno.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 115.Hodan R., Kingham K., Cotter K., Folkins A.K., Kurian A.W., Ford J.M., Longacre T. Prevalence of Lynch syndrome in women with mismatch repair-deficient ovarian cancer. Cancer Med. 2020;10:1012–1017. doi: 10.1002/cam4.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Latham A., Srinivasan P., Kemel Y., Shia J., Bandlamudi C., Mandelker D., Middha S., Hechtman J., Zehir A., Dubard-Gault M., et al. Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J. Clin. Oncol. 2019;37:286–295. doi: 10.1200/JCO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yamashita H., Nakayama K., Ishikawa M., Ishibashi T., Nakamura K., Sawada K., Yoshimura Y., Tatsumi N., Kurose S., Minamoto T., et al. Relationship between Microsatellite Instability, Immune Cells Infiltration, and Expression of Immune Checkpoint Molecules in Ovarian Carcinoma: Immunotherapeutic Strategies for the Future. Int. J. Mol. Sci. 2019;20:5129. doi: 10.3390/ijms20205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee J.-H., Cragun D., Thompson Z., Coppola M., Nicosia S.V., Akbari M., Zhang S., McLaughlin J., Narod S., Schildkraut J., et al. Association Between IHC and MSI Testing to Identify Mismatch Repair–Deficient Patients with Ovarian Cancer. Genet. Test. Mol. Biomark. 2014;18:229–235. doi: 10.1089/gtmb.2013.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosen D.G., Cai K.Q., Luthra R., Liu J. Immunohistochemical staining of hMLH1 and hMSH2 reflects microsatellite instability status in ovarian carcinoma. Mod. Pathol. 2006;19:1414–1420. doi: 10.1038/modpathol.3800672. [DOI] [PubMed] [Google Scholar]
- 120.Cai K.Q., Albarracin C., Rosen D., Zhong R., Zheng W., Luthra R., Broaddus R., Liu J. Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum. Pathol. 2004;35:552–559. doi: 10.1016/j.humpath.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 121.Travaglino A., Raffone A., Gencarelli A., Mollo A., Guida M., Insabato L., Santoro A., Zannoni G.F., Zullo F. TCGA Classification of Endometrial Cancer: The Place of Carcinosarcoma. Pathol. Oncol. Res. 2020;26:2067–2073. doi: 10.1007/s12253-020-00829-9. [DOI] [PubMed] [Google Scholar]
- 122.Cancer Genome Atlas Research Network. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bianco B., Barbosa C.P., Trevisan C.M., Laganà A.S., Montagna E. Endometrial cancer: A genetic point of view. Transl. Cancer Res. 2020;9:7706–7715. doi: 10.21037/tcr-20-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bosse T., Nout R.A., McAlpine J.N., McConechy M.K., Britton H., Hussein Y.R., Gonzalez C., Ganesan R., Steele J.C., Harrison B.T., et al. Molecular Classification of Grade 3 Endometrioid Endometrial Cancers Identifies Distinct Prognostic Subgroups. Am. J. Surg. Pathol. 2018;42:561–568. doi: 10.1097/PAS.0000000000001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pasanen A., Loukovaara M., Kaikkonen E., Olkinuora A., Pylvänäinen K., Alhopuro P., Peltomäki P., Mecklin J.P., Bützow R. Testing for Lynch Syndrome in Endometrial Carcinoma: From Universal to Age-Selective MLH1 Methylation Analysis. Cancers. 2022;14:1348. doi: 10.3390/cancers14051348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shia J., Black D., Hummer A.J., Boyd J., Soslow R.A. Routinely assessed morphological features correlate with microsatellite in-stability status in endometrial cancer. Hum. Pathol. 2008;39:116–125. doi: 10.1016/j.humpath.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 127.Mrkonjic M., Turashvili G. EPM2AIP1 Immunohistochemistry Can Be Used as Surrogate Testing for MLH1 Promoter Methyl-ation in Endometrial Cancer. Am. J. Surg. Pathol. 2022;46:376–382. doi: 10.1097/PAS.0000000000001832. [DOI] [PubMed] [Google Scholar]
- 128.Cortes-Ciriano I., Lee S., Park W.-Y., Kim T.-M., Park P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017;8:15180. doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y.K., Bashashati A., Anglesio M.S., Cochrane D.R., Grewal D.S., Ha G., McPherson A., Horlings H.M., Senz J., Prentice L.M., et al. Genomic consequences of aberrant DNA re-pair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 2017;49:856–865. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 130.Novetsky A.P., Zighelboim I., Thompson D.M., Jr., Powell M.A., Mutch D.G., Goodfellow P.J. Frequent mutations in the RPL22 gene and its clinical and functional implications. Gynecol. Oncol. 2013;128:470–474. doi: 10.1016/j.ygyno.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kawaguchi M., Banno K., Yanokura M., Kobayashi Y., Kishimi A., Ogawa S., Kisu I., Nomura H., Hirasawa A., Susumu N., et al. Analysis of candidate target genes for mononucleotide repeat mutation in microsatellite instability-high (MSI-H) endome-trial cancer. Int. J. Oncol. 2009;35:977–982. doi: 10.3892/ijo_00000411. [DOI] [PubMed] [Google Scholar]
- 132.Bilbao C., Ramírez R., Rodríguez G., Falcón O., León L., Díaz-Chico N., Perucho M., Díaz-Chico J.C. Double strand break repair components are frequent targets of microsatellite instability in endometrial cancer. Eur. J. Cancer. 2010;46:2821–2827. doi: 10.1016/j.ejca.2010.06.116. [DOI] [PubMed] [Google Scholar]
- 133.Bertoni F., Codegoni A.M., Furlan D., Tibiletti M.G., Capella C., Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosom. Cancer. 1999;26:176–180. doi: 10.1002/(SICI)1098-2264(199910)26:2<176::AID-GCC11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 134.Vassileva V., Millar A., Briollais L., Chapman W., Bapat B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002;62:4095–4099. [PubMed] [Google Scholar]
- 135.Deshpande M., Romanski P.A., Rosenwaks Z., Gerhardt J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers. 2020;12:3319. doi: 10.3390/cancers12113319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bilbao C., Rodríguez G., Ramírez R., Falcón O., León L., Chirino R., Rivero J.F., Díaz-Chico B.N., Díaz-Chico J.C., Perucho M. The relationship between microsatellite instability and PTEN gene mutations in endometrial cancer. Int. J. Cancer. 2006;119:563–570. doi: 10.1002/ijc.21862. [DOI] [PubMed] [Google Scholar]
- 137.O’Malley D.M., Bariani G.M., Cassier P.A., Marabelle A., Hansen A.R., De Jesus Acosta A., Miller W.H., Jr., Safra T., Italiano A., Mileshkin L., et al. Pembrolizumab in Patients with Microsatellite Instability–High Advanced Endometrial Cancer: Results From the KEYNOTE-158 Study. J. Clin. Oncol. 2022;40:752–761. doi: 10.1200/JCO.21.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zighelboim I., Goodfellow P.J., Gao F., Gibb R.K., Powell M.A., Rader J.S., Mutch D.G. Microsatellite instability and epigenetic inac-tivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J. Clin. Oncol. 2007;25:2042–2048. doi: 10.1200/JCO.2006.08.2107. [DOI] [PubMed] [Google Scholar]
- 139.Macdonald N.D., Salvesen H.B., Ryan A., Iversen O.E., Akslen L.A., Jacobs I.J. Frequency and prognostic impact of microsatellite instability in a large population-based study of endometrial carcinomas. Cancer Res. 2000;60:1750–1752. [PubMed] [Google Scholar]
- 140.Kanopiene D., Vidugiriene J., Valuckas K.P., Smailyte G., Uleckiene S., Bacher J. Endometrial cancer and microsatellite instability status. Open Med. 2014;10:70–76. doi: 10.1515/med-2015-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Makker V., Taylor M.H., Aghajanian C., Oaknin A., Mier J., Cohn A.L., Romeo M., Bratos R., Brose M.S., DiSimone C., et al. Lenvatinib Plus Pem-brolizumab in Patients with Advanced Endometrial Cancer. J. Clin. Oncol. 2020;38:2981–2992. doi: 10.1200/JCO.19.02627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ackroyd S.A., Huang E.S., Kurnit K.C., Lee N.K. Pembrolizumab and lenvatinib versus carboplatin and paclitaxel as first-line therapy for advanced or recurrent endometrial cancer: A Markov analysis. Gynecol. Oncol. 2021;162:249–255. doi: 10.1016/j.ygyno.2021.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Oaknin A., Gilbert L., Tinker A.V., Brown J., Mathews C., Press J., Sabatier R., O’Malley D.M., Samouelian V., Boni V., et al. Safety and antitumor activity of dostarlimab in patients with advanced or recurrent DNA mismatch repair deficient/microsatellite insta-bility-high (dMMR/MSI-H) or proficient/stable (MMRp/MSS) endometrial cancer: Interim results from GARNET-a phase I, single-arm study. Immunother Cancer. 2022;10:e003777. doi: 10.1136/jitc-2021-003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ukkola I., Nummela P., Pasanen A., Kero M., Lepistö A., Kytölä S., Bützow R., Ristimäki A. Detection of microsatellite instability with Idylla MSI assay in colorectal and endometrial cancer. Virchows Arch. 2021;479:471–479. doi: 10.1007/s00428-021-03082-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saeki H., Hlaing M.T., Horimoto Y., Kajino K., Ohtsuji N., Fujino K., Terao Y., Hino O. Usefulness of immunohistochemistry for mismatch repair protein and microsatellite instability examination in adenocarcinoma and background endometrium of spo-radic endometrial cancer cases. J. Obs. Gynaecol. Res. 2019;45:2037–2042. doi: 10.1111/jog.14061. [DOI] [PubMed] [Google Scholar]
- 146.Siemanowski J., Schömig-Markiefka B., Buhl T., Haak A., Siebolts U., Dietmaier W., Arens N., Pauly N., Ataseven B., Büttner R., et al. Managing Difficulties of Microsatellite Instability Testing in Endometrial Cancer-Limitations and Ad-vantages of Four Different PCR-Based Approaches. Cancers. 2021;13:1268. doi: 10.3390/cancers13061268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McConechy M., Talhouk A., Li-Chang H., Leung S., Huntsman D., Gilks C., McAlpine J. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol. Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 148.Song Y., Gu Y., Hu X., Wang M., He Q., Li Y. Endometrial Tumors with MSI-H and dMMR Share a Similar Tumor Immune Mi-croenvironment. Onco Targets Ther. 2021;14:4485–4497. doi: 10.2147/OTT.S324641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Warth A., Körner S., Penzel R., Muley T., Dienemann H., Schirmacher P., von Knebel-Doeberitz M., Weichert W., Kloor M. Mi-crosatellite instability in pulmonary adenocarcinomas: A comprehensive study of 480 cases. Virchows Arch. 2016;468:313–319. doi: 10.1007/s00428-015-1892-7. [DOI] [PubMed] [Google Scholar]
- 150.De Marchi P., Berardinelli G.N., Cavagna R., De Paula F., Da Silva E.A., Miziara J., Leal L., Reis R. EP1.04-11 Frequency of Microsatellite Instability (MSI) in Brazilian TKI non-treatable Non-Small Cell Lung Cancer (NSCLC) patients. J. Thor. Oncol. 2019;14:S973. doi: 10.1016/j.jtho.2019.08.2133. [DOI] [Google Scholar]
- 151.Woenckhaus M., Stoehr R., Dietmaier W., Wild P.J., Zieglmeier U., Foerster J., Merk J., Blaszyk H., Pfeifer M., Hofstaedter F., et al. Microsatellite instability at chromosome 8p in non-small cell lung cancer is associated with lymph node metastasis and squamous differentiation. Int. J. Oncol. 2003;23:1357–1363. doi: 10.3892/ijo.23.5.1357. [DOI] [PubMed] [Google Scholar]
- 152.Chang J.W., Chen Y.C., Chen C.Y., Chen J.T., Chen S.K., Wang Y.C. Correlation of genetic instability with mismatch repair protein expression and p53 mutations in non-small cell lung cancer. Clin. Cancer Res. 2000;6:1639–1646. [PubMed] [Google Scholar]
- 153.Carpagnano G.E., Lacedonia D., Crisetti E., Martinelli D., Foschino-Barbaro M.P. New panel of microsatellite alterations detecta-ble in the EBC for lung cancer prognosis. J. Cancer. 2016;7:2266–2269. doi: 10.7150/jca.15921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olivares-Hernández A., Del Barco Morillo E., Parra Pérez C., Miramontes-González J.P., Figuero-Pérez L., Martín-Gómez T., Escala-Cornejo R., Bellido Hernández L., González Sarmiento R., Cruz-Hernández J.J., et al. Influence of DNA Mismatch Repair (MMR) System in Survival and Response to Immune Checkpoint Inhibitors (ICIs) in Non-Small Cell Lung Cancer (NSCLC): Retrospective Analysis. Biomedicines. 2022;10:360. doi: 10.3390/biomedicines10020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Leighl N.B., Hellmann M.D., Hui R., Carcereny E., Felip E., Ahn M.-J., Eder J.P., Balmanoukian A.S., Aggarwal C., Horn L., et al. Pembrolizumab in patients with advanced non-small-cell lung cancer (KEYNOTE-001): 3-year results from an open-label, phase 1 study. Lancet Respir. Med. 2019;7:347–357. doi: 10.1016/S2213-2600(18)30500-9. [DOI] [PubMed] [Google Scholar]
- 156.Shaverdian N., Lisberg A.E., Bornazyan K., Veruttipong D., Goldman J.W., Formenti S.C., Garon E.B., Lee P. Previous radiother-apy and the clinical activity and toxicity of pembrolizumab in the treatment of non-small-cell lung cancer: A secondary analy-sis of the KEYNOTE-001 phase 1 trial. Lancet Oncol. 2017;18:895–903. doi: 10.1016/S1470-2045(17)30380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Diaz L.A., Marabelle A., Delord J.-P., Shapira-Frommer R., Geva R., Peled N., Kim T.W., Andre T., Van Cutsem E., Guimbaud R., et al. Pembrolizumab therapy for mi-crosatellite instability high (MSI-H) colorectal cancer (CRC) and non-CRC. J. Clin. Oncol. 2017;35:3071. doi: 10.1200/JCO.2017.35.15_suppl.3071. [DOI] [Google Scholar]
- 158.Mills A.M., Dill E.A., Moskaluk C.A., Dziegielewski J., Bullock T.N., Dillon P.M. The Relationship Between Mismatch Repair Defi-ciency and PD-L1 Expression in Breast Carcinoma. Am. J. Surg. Pathol. 2018;42:183–191. doi: 10.1097/PAS.0000000000000949. [DOI] [PubMed] [Google Scholar]
- 159.Sajjadi E., Venetis K., Piciotti R., Invernizzi M., Guerini-Rocco E., Haricharan S., Fusco N. Mismatch repair-deficient hormone receptor-positive breast cancers: Biology and pathological characterization. Cancer Cell Int. 2021;21:266. doi: 10.1186/s12935-021-01976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fusco N., Lopez G., Corti C., Pesenti C., Colapietro P., Ercoli G., Gaudioso G., Faversani A., Gambini D., Michelotti A., et al. Mismatch Repair Protein Loss as a Prognostic and Predictive Biomarker in Breast Cancers Regardless of Microsatellite Instability. JNCI Cancer Spectr. 2018;2:pky056. doi: 10.1093/jncics/pky056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lotsari J.E., Gylling A., Abdel-Rahman W.M., Nieminen T.T., Aittomäki K., Friman M., Pitkänen R., Aarnio M., Järvinen H.J., Mecklin J.P., et al. Breast carcinoma and Lynch syndrome: Molecular analysis of tumors arising in mutation carriers, non-carriers, and sporadic cases. Breast Cancer Res. 2012;14:R90. doi: 10.1186/bcr3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kurata K., Kubo M., Kai M., Mori H., Kawaji H., Kaneshiro K., Yamada M., Nishimura R., Osako T., Arima N., et al. Microsatellite instability in Japanese female patients with triple-negative breast cancer. Breast Cancer. 2020;27:490–498. doi: 10.1007/s12282-019-01043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Caldés T., Perez-Segura P., Tosar A., de La Hoya M., Diaz-Rubio E. Microsatellite instability correlates with negative expression of estrogen and progesterone receptors in sporadic breast cancer. Teratog. Carcinog. Mutagen. 2000;20:283–291. doi: 10.1002/1520-6866(2000)20:5<283::AID-TCM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 164.Yee C.J., Roodi N., Verrier C.S., Parl F.F. Microsatellite instability and loss of heterozygosity in breast cancer. Cancer Res. 1994;54:1641–1644. [PubMed] [Google Scholar]
- 165.Rugo H.S., Delord J.-P., Im S.-A., Ott P.A., Piha-Paul S.A., Bedard P.L., Sachdev J., Le Tourneau C., van Brummelen E.M., Varga A., et al. Safety and Antitumor Activity of Pembrolizumab in Patients with Estrogen Receptor–Positive/Human Epidermal Growth Factor Receptor 2–Negative Advanced Breast Cancer. Clin. Cancer Res. 2018;24:2804–2811. doi: 10.1158/1078-0432.CCR-17-3452. [DOI] [PubMed] [Google Scholar]
- 166.Lupinacci R.M., Goloudina A., Buhard O., Bachet J.B., Maréchal R., Demetter P., Cros J., Bardier-Dupas A., Collura A., Cervera P., et al. Preva-lence of Microsatellite Instability in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology. 2018;154:1061–1065. doi: 10.1053/j.gastro.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 167.Leclerc J., Vermaut C., Buisine M.-P. Diagnosis of Lynch Syndrome and Strategies to Distinguish Lynch-Related Tumors from Sporadic MSI/dMMR Tumors. Cancers. 2021;13:467. doi: 10.3390/cancers13030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hu Z.I., Shia J., Stadler Z.K., Varghese A.M., Capanu M., Salo-Mullen E., Lowery M.A., Diaz L.A., Jr., Mandelker D., Yu K.H., et al. Evaluating Mismatch Repair Deficiency in Pancreatic Adenocarcinoma: Challenges and Recommendations. Clin. Cancer Res. 2018;24:1326–1336. doi: 10.1158/1078-0432.CCR-17-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Møller P., Seppälä T.T., Bernstein I., Holinski-Feder E., Sala P., Gareth Evans D., Lindblom A., Macrae F., Blanco I., Sijmons R.H., et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut. 2018;67:1306–1316. doi: 10.1136/gutjnl-2017-314057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ahmad-Nielsen S.A., Nielsen M.F.B., Mortensen M.B., Detlefsen S. Frequency of mismatch repair deficiency in pancreatic ductal adenocarcinoma. Pathol. Res. Pract. 2020;216:152985. doi: 10.1016/j.prp.2020.152985. [DOI] [PubMed] [Google Scholar]
- 171.Lupinacci R.M., Bachet J.B., André T., Duval A., Svrcek M. Pancreatic ductal adenocarcinoma harboring microsatellite instability/DNA mismatch repair deficiency. Towards personalized medicine. Surg. Oncol. 2019;28:121–127. doi: 10.1016/j.suronc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 172.Riazy M., Kalloger S.E., Sheffield B.S., Peixoto R.D., Li-Chang H.H., Scudamore C.H., Renouf D.J., Schaeffer D.F. Mismatch repair status may predict response to adjuvant chemotherapy in resectable pancreatic ductal adenocarcinoma. Mod. Pathol. 2015;28:1383–1389. doi: 10.1038/modpathol.2015.89. [DOI] [PubMed] [Google Scholar]
- 173.Cloyd J.M., Katz M.H.G., Wang H., Cuddy A., You Y.N. Clinical and Genetic Implications of DNA Mismatch Repair Deficiency in Patients with Pancreatic Ductal Adenocarcinoma. JAMA Surg. 2017;152:1086–1088. doi: 10.1001/jamasurg.2017.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Marabelle A., Le D.T., Ascierto P.A., Di Giacomo A.M., De Jesus-Acosta A., Delord J.P., Geva R., Gottfried M., Penel N., Hansen A.R., et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Can-cer: Results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu B., Fuchs T., Dogan S., Landa I., Katabi N., Fagin J.A., Tuttle R.M., Sherman E., Gill A.J., Ghossein R. Dissecting Anaplastic Thy-roid Carcinoma: A Comprehensive Clinical, Histologic, Immunophenotypic, and Molecular Study of 360 Cases. Thyroid. 2020;30:1505–1517. doi: 10.1089/thy.2020.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wong K.S., Lorch J.H., Alexander E.K., Nehs M.A., Nowak J.A., Hornick J.L., Barletta J.A. Clinicopathologic Features of Mismatch Repair-Deficient Anaplastic Thyroid Carcinomas. Thyroid. 2019;29:666–673. doi: 10.1089/thy.2018.0716. [DOI] [PubMed] [Google Scholar]
- 177.Qiao P.-P., Tian K.-S., Han L.-T., Ma B., Shen C.-K., Zhao R.-Y., Zhang Y., Wei W.-J., Chen X.-P. Correlation of mismatch repair deficiency with clinicopathological features and programmed death-ligand 1 expression in thyroid carcinoma. Endocrine. 2022;76:660–670. doi: 10.1007/s12020-022-03031-w. [DOI] [PubMed] [Google Scholar]
- 178.D’Andréa G., Lassalle S., Guevara N., Mograbi B., Hofman P. From biomarkers to therapeutic targets: The promise of PD-L1 in thyroid autoimmunity and cancer. Theranostics. 2021;11:1310–1325. doi: 10.7150/thno.50333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rocha M.L., Schmid K.W., Czapiewski P. The prevalence of DNA microsatellite instability in anaplastic thyroid carcinoma–systematic review and discussion of current therapeutic options. Contemp. Oncol. 2021;25:213–223. doi: 10.5114/wo.2021.110052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ülgen E., Can Ö., Bilguvar K., Oktay Y., Akyerli C.B., Danyeli A.E., Yakıcıer M.C., Sezerman O.U., Pamir M.N., Özduman K. Whole exome sequencing-based analysis to identify DNA damage repair deficiency as a major contributor to gliomagenesis in adult diffuse gliomas. J. Neurosurg. 2019;132:1435–1446. doi: 10.3171/2019.1.JNS182938. [DOI] [PubMed] [Google Scholar]
- 181.Akır E., Saygın İ., Ercin M.E. Investigation of the relationship between immune checkpoints and mismatch repair deficiency in recurrent and nonrecurrent glioblastoma. Turk. J. Med. Sci. 2021;51:1800–1808. doi: 10.3906/sag-2010-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kroiss M.M., Vogt T.M.M., Landthaler M., Schlegel J., Stolz W. Microsatellite Instability in Malignant Melanomas. Acta Derm. Venereol. 2001;81:242–245. doi: 10.1080/00015550152572840. [DOI] [PubMed] [Google Scholar]
- 183.Birindelli S., Tragni G., Bartoli C., Ranzani G.N., Rilke F., Pierotti M.A., Pilotti S. Detection of microsatellite alterations in the spec-trum of melanocytic nevi in patients with or without individual or family history of melanoma. Int. J. Cancer. 2000;86:255–261. doi: 10.1002/(SICI)1097-0215(20000415)86:2<255::AID-IJC16>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 184.Uribe P., Wistuba I.I., Gonzalez S. Allelotyping, microsatellite instability, and BRAF mutation analyses in common and atypical melanocytic nevi and primary cutaneous melanomas. Am. J. Dermatopathol. 2009;31:354–363. doi: 10.1097/DAD.0b013e318185d205. [DOI] [PubMed] [Google Scholar]
- 185.Kubecek O., Trojanova P., Molnarova V., Kopecky J. Microsatellite instability as a predictive factor for immunotherapy in ma-lignant melanoma. Med. Hypotheses. 2016;93:74–76. doi: 10.1016/j.mehy.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 186.Gayhart M.G., Johnson N., Paul A., Quillin J.M., Hampton L.J., Idowu M.O., Smith S.C. Universal Mismatch Repair Protein Screening in Upper Tract Urothelial Carcinoma. Am. J. Clin. Pathol. 2020;154:792–801. doi: 10.1093/ajcp/aqaa100. [DOI] [PubMed] [Google Scholar]
- 187.Lindner A.K., Schachtner G., Tulchiner G., Thurnher M., Untergasser G., Obrist P., Pipp I., Steinkohl F., Horninger W., Culig Z., et al. Lynch Syndrome: Its Impact on Urothelial Carcinoma. Int. J. Mol. Sci. 2021;22:531. doi: 10.3390/ijms22020531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fraune C., Simon R., Hube-Magg C., Makrypidi-Fraune G., Kähler C., Kluth M., Höflmayer D., Büscheck F., Dum D., Luebke A.M., et al. MMR deficiency in urothelial carcinoma of the bladder presents with temporal and spatial homogeneity throughout the tumor mass. Urol. Oncol. Semin. Orig. Investig. 2020;38:488–495. doi: 10.1016/j.urolonc.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 189.Metcalfe M.J., Petros F.G., Rao P., Mork M.E., Xiao L., Broaddus R.R., Matin S.F. Universal Point of Care Testing for Lynch Syn-drome in Patients with Upper Tract Urothelial Carcinoma. J. Urol. 2018;199:60–65. doi: 10.1016/j.juro.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 190.Harper H.L., McKenney J.K., Heald B., Stephenson A., Campbell S.C., Plesec T., Magi-Galluzzi C. Upper tract urothelial carcino-mas: Frequency of association with mismatch repair protein loss and lynch syndrome. Mod. Pathol. 2017;30:146–156. doi: 10.1038/modpathol.2016.171. [DOI] [PubMed] [Google Scholar]
- 191.Urakami S., Inoshita N., Oka S., Miyama Y., Nomura S., Arai M., Sakaguchi K., Kurosawa K., Okaneya T. Clinicopathological characteristics of patients with upper urinary tract urothelial cancer with loss of immunohistochemical expression of the DNA mismatch repair proteins in universal screening. Int. J. Urol. 2017;25:151–156. doi: 10.1111/iju.13481. [DOI] [PubMed] [Google Scholar]
- 192.Ju J., Mills A.M., Mahadevan M.S., Fan J., Culp S.H., Thomas M.H., Cathro H.P. Universal Lynch Syndrome Screening Should be Performed in All Upper Tract Urothelial Carcinomas. Am. J. Surg. Pathol. 2018;42:1549–1555. doi: 10.1097/PAS.0000000000001141. [DOI] [PubMed] [Google Scholar]
- 193.Guedes L.B., Antonarakis E.S., Schweizer M.T., Mirkheshti N., Almutairi F., Park J.C., Glavaris S., Hicks J., Eisenberger M.A., De Marzo A.M., et al. MSH2 Loss in Primary Prostate Cancer. Clin. Cancer Res. 2017;23:6863–6874. doi: 10.1158/1078-0432.CCR-17-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sedhom R., Antonarakis E.S. Clinical implications of mismatch repair deficiency in prostate cancer. Futur. Oncol. 2019;15:2395–2411. doi: 10.2217/fon-2019-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mollica V., Marchetti A., Rosellini M., Nuvola G., Rizzo A., Santoni M., Cimadamore A., Montironi R., Massari F. An Insight on Novel Molecular Pathways in Metastatic Prostate Cancer: A Focus on DDR, MSI and AKT. Int. J. Mol. Sci. 2021;22:13519. doi: 10.3390/ijms222413519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.